6RLP
 
 | | Cryo-EM reconstruction of TMV coat protein | | Descriptor: | Capsid protein, RNA (5'-R(P*GP*AP*A)-3') | | Authors: | Kandiah, E, Effantin, G. | | Deposit date: | 2019-05-02 | | Release date: | 2022-11-23 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | CM01: a facility for cryo-electron microscopy at the European Synchrotron.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
5MKC
 
 | | Crystal structure of the RrgA Jo.In complex | | Descriptor: | CALCIUM ION, Cell wall surface anchor family protein (Jo),Cell wall surface anchor family protein (In), NICKEL (II) ION, ... | | Authors: | Bonnet, J, Cartannaz, J, Tourcier, G, Contreras-Martel, C, Kleman, J.P, Fenel, D, Schoehn, G, Morlot, C, Vernet, T, Di Guilmi, A.M. | | Deposit date: | 2016-12-03 | | Release date: | 2017-03-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Autocatalytic association of proteins by covalent bond formation: a Bio Molecular Welding toolbox derived from a bacterial adhesin.
Sci Rep, 7, 2017
|
|
6GFJ
 
 | | Structure of RIP2 CARD domain fused to crystallisable MBP tag | | Descriptor: | Sugar ABC transporter substrate-binding protein,Receptor-interacting serine/threonine-protein kinase 2, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Pellegrini, E, Cusack, S. | | Deposit date: | 2018-04-30 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | RIP2 filament formation is required for NOD2 dependent NF-kappa B signalling.
Nat Commun, 9, 2018
|
|
5LYE
 
 | |
1AN1
 
 | | LEECH-DERIVED TRYPTASE INHIBITOR/TRYPSIN COMPLEX | | Descriptor: | CALCIUM ION, TRYPSIN, TRYPTASE INHIBITOR | | Authors: | Priestle, J.P, Di Marco, S. | | Deposit date: | 1997-06-26 | | Release date: | 1998-07-01 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structure of the complex of leech-derived tryptase inhibitor (LDTI) with trypsin and modeling of the LDTI-tryptase system.
Structure, 5, 1997
|
|
5X0N
 
 | |
5X0O
 
 | |
6LVV
 
 | | N, N-dimethylformamidase | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, N,N-dimethylformamidase large subunit, ... | | Authors: | Arya, C.K, Ramaswamy, S, Kutti, R.V, Gurunath, R. | | Deposit date: | 2020-02-05 | | Release date: | 2020-08-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A 2-Tyr-1-carboxylate Mononuclear Iron Center Forms the Active Site of a Paracoccus Dimethylformamidase.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
5CPR
 
 | | The novel SUV4-20 inhibitor A-196 verifies a role for epigenetics in genomic integrity | | Descriptor: | 6,7-dichloro-N-cyclopentyl-4-(pyridin-4-yl)phthalazin-1-amine, Histone-lysine N-methyltransferase SUV420H1, S-ADENOSYLMETHIONINE, ... | | Authors: | Jakob, C.G, Upadhyay, A.K, Sun, C. | | Deposit date: | 2015-07-21 | | Release date: | 2017-01-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | The SUV4-20 inhibitor A-196 verifies a role for epigenetics in genomic integrity.
Nat. Chem. Biol., 13, 2017
|
|
2QRM
 
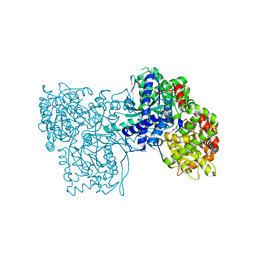 | | Glycogen Phosphorylase b in complex with (1R)-3'-(4-nitrophenyl)-spiro[1,5-anhydro-D-glucitol-1,5'-isoxazoline] | | Descriptor: | (3S,5R,7R,8S,9S,10R)-7-(hydroxymethyl)-3-(4-nitrophenyl)-1,6-dioxa-2-azaspiro[4.5]decane-8,9,10-triol, Glycogen phosphorylase, muscle form | | Authors: | Gizilis, G, Alexacou, K.M, Chrysina, E.D, Zographos, S.E, Leonidas, D.D, Oikonomakos, N.G. | | Deposit date: | 2007-07-28 | | Release date: | 2008-07-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Glucose-based spiro-isoxazolines: a new family of potent glycogen phosphorylase inhibitors.
Bioorg.Med.Chem., 17, 2009
|
|
2QRP
 
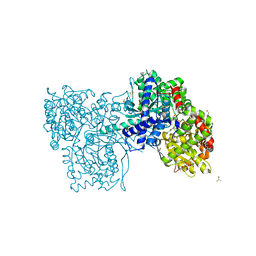 | | Glycogen Phosphorylase b in complex with (1R)-3'-(2-naphthyl)-spiro[1,5-anhydro-D-glucitol-1,5'-isoxazoline] | | Descriptor: | (3S,5R,7R,8S,9S,10R)-7-(hydroxymethyl)-3-(2-naphthyl)-1,6-dioxa-2-azaspiro[4.5]decane-8,9,10-triol, DIMETHYL SULFOXIDE, Glycogen phosphorylase, ... | | Authors: | Gizilis, G, Alexacou, K.M, Chrysina, E.D, Zographos, S.E, Leonidas, D.D, Oikonomakos, N.G. | | Deposit date: | 2007-07-28 | | Release date: | 2008-07-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Glucose-based spiro-isoxazolines: a new family of potent glycogen phosphorylase inhibitors.
Bioorg.Med.Chem., 17, 2009
|
|
2QRQ
 
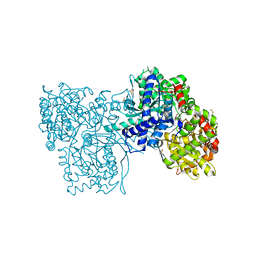 | | Glycogen Phosphorylase b in complex with (1R)-3'-(4-methylphenyl)-spiro[1,5-anhydro-D-glucitol-1,5'-isoxazoline] | | Descriptor: | (3S,5R,7R,8S,9S,10R)-7-(hydroxymethyl)-3-(4-methylphenyl)-1,6-dioxa-2-azaspiro[4.5]decane-8,9,10-triol, DIMETHYL SULFOXIDE, Glycogen phosphorylase, ... | | Authors: | Gizilis, G, Alexacou, K.M, Chrysina, E.D, Zographos, S.E, Leonidas, D.D, Oikonomakos, N.G. | | Deposit date: | 2007-07-28 | | Release date: | 2008-07-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Glucose-based spiro-isoxazolines: a new family of potent glycogen phosphorylase inhibitors.
Bioorg.Med.Chem., 17, 2009
|
|
6R81
 
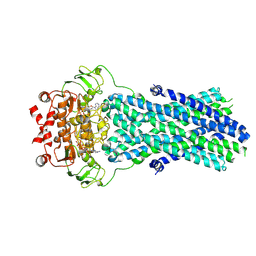 | | Multidrug resistance transporter BmrA mutant E504A bound with ATP and Mg solved by Cryo-EM | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Lipid A export ATP-binding/permease protein MsbA, MAGNESIUM ION | | Authors: | Wiseman, B, Chaptal, V, Zampieri, V, Magnard, S, Hogbom, M, Falson, P. | | Deposit date: | 2019-03-30 | | Release date: | 2020-05-06 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Substrate-bound and substrate-free outward-facing structures of a multidrug ABC exporter.
Sci Adv, 8, 2022
|
|
6R72
 
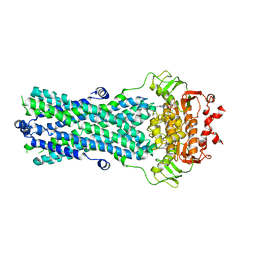 | | Crystal structure of BmrA-E504A in an outward-facing conformation | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Multidrug exporter ATP-binding cassette | | Authors: | Chaptal, V, Zampieri, V, Kilburg, A, Magnard, S, Falson, P. | | Deposit date: | 2019-03-28 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.95 Å) | | Cite: | Substrate-bound and substrate-free outward-facing structures of a multidrug ABC exporter.
Sci Adv, 8, 2022
|
|
6ZLV
 
 | | MreC | | Descriptor: | Rod shape-determining protein MreC | | Authors: | Estrozi, L.F, Contreras-Martel, C. | | Deposit date: | 2020-07-01 | | Release date: | 2021-03-17 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Self-association of MreC as a regulatory signal in bacterial cell wall elongation.
Nat Commun, 12, 2021
|
|
6ZM0
 
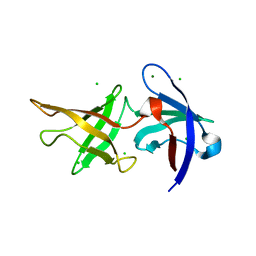 | |
2QRH
 
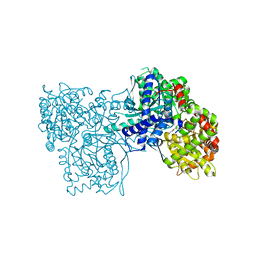 | | Glycogen Phosphorylase b in complex with (1R)-3'-phenylspiro[1,5-anhydro-D-glucitol-1,5'-isoxazoline] | | Descriptor: | (5R,7R,8S,9S,10R)-7-(hydroxymethyl)-3-phenyl-1,6-dioxa-2-azaspiro[4.5]dec-2-ene-8,9,10-triol, Glycogen phosphorylase, muscle form | | Authors: | Kizilis, G, Alexacou, K.-M, Chrysina, E.D, Zographos, S.E, Leonidas, D.D, Oikonomakos, N.G. | | Deposit date: | 2007-07-28 | | Release date: | 2008-08-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Glucose-based spiro-isoxazolines: a new family of potent glycogen phosphorylase inhibitors.
Bioorg.Med.Chem., 17, 2009
|
|
6Y1Z
 
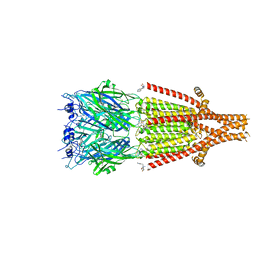 | | Mouse serotonin 5HT3 receptor in complex with palonosetron | | Descriptor: | (3~{a}~{S})-2-[(3~{S})-1-azabicyclo[2.2.2]octan-3-yl]-3~{a},4,5,6-tetrahydro-3~{H}-benzo[de]isoquinolin-1-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zarkadas, E, Perot, J, Nury, H. | | Deposit date: | 2020-02-14 | | Release date: | 2020-03-04 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | The Binding of Palonosetron and Other Antiemetic Drugs to the Serotonin 5-HT3 Receptor.
Structure, 28, 2020
|
|
7PCD
 
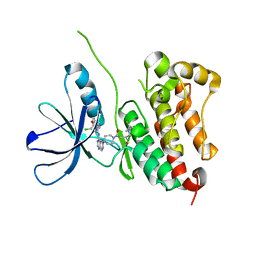 | | HER2 IN COMPLEX WITH A COVALENT INHIBITOR | | Descriptor: | 1-[4-[4-[[3,5-bis(chloranyl)-4-([1,2,4]triazolo[1,5-a]pyridin-7-yloxy)phenyl]amino]pyrimido[5,4-d]pyrimidin-6-yl]piperazin-1-yl]-4-(3-fluoranylazetidin-1-yl)butan-1-one, Receptor tyrosine-protein kinase erbB-2 | | Authors: | Bader, G. | | Deposit date: | 2021-08-03 | | Release date: | 2022-07-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Discovery of potent and selective HER2 inhibitors with efficacy against HER2 exon 20 insertion-driven tumors, which preserve wild-type EGFR signaling.
Nat Cancer, 3, 2022
|
|
8IWQ
 
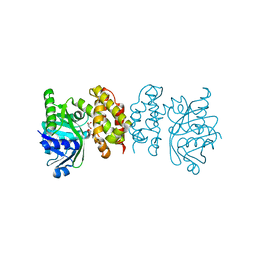 | |
8IXM
 
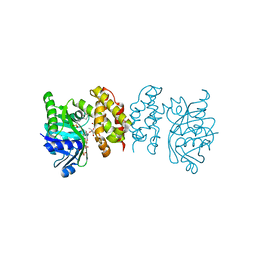 | |
8IWG
 
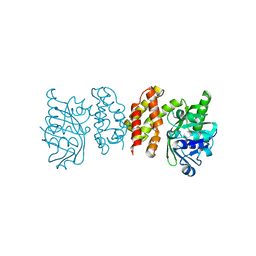 | |
8IXH
 
 | |
8IX9
 
 | | Pseudomoans Aerugiona Wildtype Ketopantoate Reductase with NADPH | | Descriptor: | 2-dehydropantoate 2-reductase, FORMIC ACID, GLYCEROL, ... | | Authors: | Choudhury, G.B, Datta, S. | | Deposit date: | 2023-03-31 | | Release date: | 2024-04-03 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Implication of Molecular Constraints Facilitating the Functional Evolution of Pseudomonas aeruginosa KPR2 into a Versatile alpha-Keto-Acid Reductase.
Biochemistry, 63, 2024
|
|
2QRG
 
 | | Glycogen Phosphorylase b in complex with (1R)-3'-(4-methoxyphenyl)-spiro[1,5-anhydro-D-glucitol-1,5'-isoxazoline] | | Descriptor: | (5R,7R,8S,9S,10R)-7-(HYDROXYMETHYL)-3-(4-METHOXYPHENYL)-1,6-DIOXA-2-AZASPIRO[4.5]DEC-2-ENE-8,9,10-TRIOL, Glycogen phosphorylase, muscle form | | Authors: | Kizilis, G, Alexacou, K.-M, Chrysina, E.D, Zographos, S.E, Leonidas, D.D, Oikonomakos, N.G. | | Deposit date: | 2007-07-28 | | Release date: | 2008-08-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Glucose-based spiro-isoxazolines: a new family of potent glycogen phosphorylase inhibitors.
Bioorg.Med.Chem., 17, 2009
|
|
