8FCZ
 
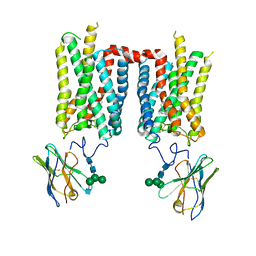 | | Crystal structure of ground-state rhodopsin in complex with a nanobody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody Nb2, ... | | Authors: | Salom, D, Palczewski, K, Kiser, P.D. | | Deposit date: | 2022-12-01 | | Release date: | 2023-08-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural basis for the allosteric modulation of rhodopsin by nanobody binding to its extracellular domain.
Nat Commun, 14, 2023
|
|
8RS0
 
 | |
8RRS
 
 | | Structure of mouse RyR2 solubilised in detergent in open state in complex with Ca2+, ATP, caffeine and Nb9657. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CAFFEINE, CALCIUM ION, ... | | Authors: | Li, C, Efremov, R.G. | | Deposit date: | 2024-01-23 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Rapid small-scale nanobody-assisted purification of ryanodine receptors for cryo-EM.
J.Biol.Chem., 300, 2024
|
|
8RRT
 
 | |
8RRU
 
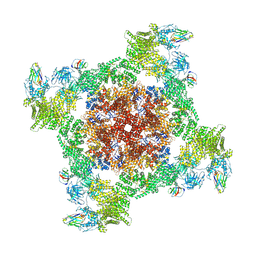 | |
8RRW
 
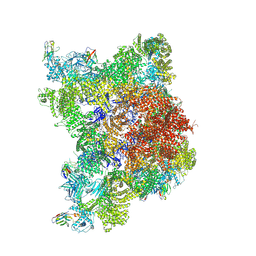 | |
8RRV
 
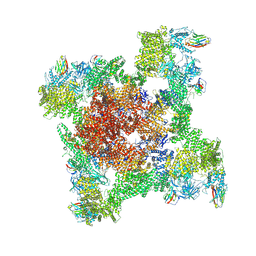 | |
8S5Z
 
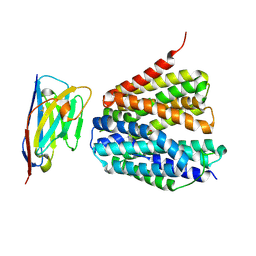 | |
8S62
 
 | |
8S4U
 
 | |
8S5U
 
 | | Cryo-EM structure of thiamine-bound human SLC19A3 in outward-open state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(4-AMINO-2-METHYL-PYRIMIDIN-5-YLMETHYL)-5-(2-HYDROXY-ETHYL)-4-METHYL-THIAZOL-3-IUM, Nb3.4, ... | | Authors: | Gabriel, F, Loew, C. | | Deposit date: | 2024-02-26 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Structural basis of thiamine transport and drug recognition by SLC19A3.
Nat Commun, 15, 2024
|
|
8S61
 
 | |
8S5W
 
 | | Cryo-EM structure of fedratinib-bound human SLC19A3 in inward-open state | | Descriptor: | N-tert-butyl-3-{[5-methyl-2-({4-[2-(pyrrolidin-1-yl)ethoxy]phenyl}amino)pyrimidin-4-yl]amino}benzenesulfonamide, Nb3.7, Thiamine transporter 2 | | Authors: | Gabriel, F, Loew, C. | | Deposit date: | 2024-02-26 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Structural basis of thiamine transport and drug recognition by SLC19A3.
Nat Commun, 15, 2024
|
|
8RRX
 
 | | Structure of RyR1 reconstituted into lipid nanodisc in primed state in complex with Ca2+, ATP, caffeine and Nb9657 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, ADENOSINE-5'-TRIPHOSPHATE, CAFFEINE, ... | | Authors: | Li, C, Efremov, R.G. | | Deposit date: | 2024-01-23 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Rapid small-scale nanobody-assisted purification of ryanodine receptors for cryo-EM.
J.Biol.Chem., 300, 2024
|
|
5GSP
 
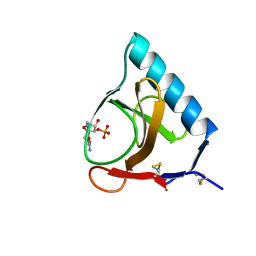 | | RIBONUCLEASE T1/3'-GMP, 9 WEEKS | | Descriptor: | CALCIUM ION, GUANOSINE-3'-MONOPHOSPHATE, RIBONUCLEASE T1 | | Authors: | Zegers, I, Wyns, L. | | Deposit date: | 1997-12-09 | | Release date: | 1998-03-18 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Hydrolysis of a slow cyclic thiophosphate substrate of RNase T1 analyzed by time-resolved crystallography.
Nat.Struct.Biol., 5, 1998
|
|
5G5X
 
 | |
5G5R
 
 | |
6N50
 
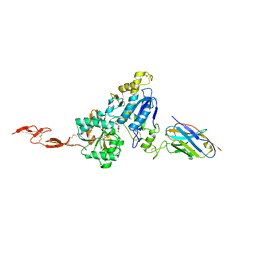 | | Metabotropic Glutamate Receptor 5 Extracellular Domain in Complex with Nb43 and L-quisqualic acid | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 5, ... | | Authors: | Koehl, A, Hu, H, Feng, D, Sun, B, Chu, M, Weis, W.I, Skiniotis, G, Mathiesen, J.M, Kobilka, B.K. | | Deposit date: | 2018-11-20 | | Release date: | 2019-01-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.751 Å) | | Cite: | Structural insights into the activation of metabotropic glutamate receptors.
Nature, 566, 2019
|
|
4AQ1
 
 | |
6N4Y
 
 | | Metabotropic Glutamate Receptor 5 Extracellular Domain with Nb43 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 5, ... | | Authors: | Koehl, A, Hu, H, Feng, D, Sun, B, Chu, M, Weis, W.I, Mathiesen, J.M, Skiniotis, G, Kobilka, B.K. | | Deposit date: | 2018-11-20 | | Release date: | 2019-01-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.262 Å) | | Cite: | Structural insights into the activation of metabotropic glutamate receptors.
Nature, 566, 2019
|
|
6N4X
 
 | | Metabotropic Glutamate Receptor 5 Apo Form Ligand Binding Domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, Metabotropic glutamate receptor 5 | | Authors: | Koehl, A, Hu, H, Feng, D, Sun, B, Weis, W.I, Skiniotis, G.S, Mathiesen, J.M, Kobilka, B.K. | | Deposit date: | 2018-11-20 | | Release date: | 2019-01-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structural insights into the activation of metabotropic glutamate receptors.
Nature, 566, 2019
|
|
6N52
 
 | | Metabotropic Glutamate Receptor 5 Apo Form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 5 | | Authors: | Koehl, A, Hu, H, Feng, D, Sun, B, Weis, W.I, Skiniotis, G.S, Mathiesen, J.M, Kobilka, B.K. | | Deposit date: | 2018-11-20 | | Release date: | 2019-01-23 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural insights into the activation of metabotropic glutamate receptors.
Nature, 566, 2019
|
|
6N51
 
 | | Metabotropic Glutamate Receptor 5 bound to L-quisqualate and Nb43 | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 5, ... | | Authors: | Koehl, A, Hu, H, Feng, D, Sun, B, Weis, W.I, Skiniotis, G.S, Mathiesen, J.M, Kobilka, B.K. | | Deposit date: | 2018-11-20 | | Release date: | 2019-01-23 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural insights into the activation of metabotropic glutamate receptors.
Nature, 566, 2019
|
|
6NI2
 
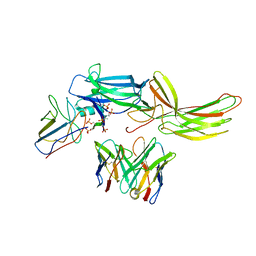 | | Stabilized beta-arrestin 1-V2T subcomplex of a GPCR-G protein-beta-arrestin mega-complex | | Descriptor: | Beta-arrestin-1, Fab30 Heavy Chain, Fab30 Light Chain, ... | | Authors: | Nguyen, A.H, Thomsen, A.R.B, Cahill, T.J, des Georges, A, Lefkowitz, R.J. | | Deposit date: | 2018-12-26 | | Release date: | 2019-11-20 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of an endosomal signaling GPCR-G protein-beta-arrestin megacomplex.
Nat.Struct.Mol.Biol., 26, 2019
|
|
5USF
 
 | |
