8JY4
 
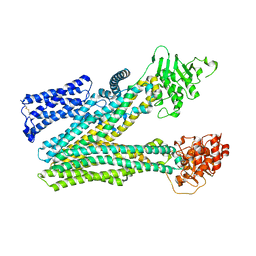 | | Cryo-EM structure of human ABC transporter ABCC2 in apo' state | | Descriptor: | ATP-binding cassette sub-family C member 2 | | Authors: | Mao, Y.X, Chen, Z.P, Wang, L, Hou, W.T, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2023-07-02 | | Release date: | 2024-01-03 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Transport mechanism of human bilirubin transporter ABCC2 tuned by the inter-module regulatory domain.
Nat Commun, 15, 2024
|
|
8JXU
 
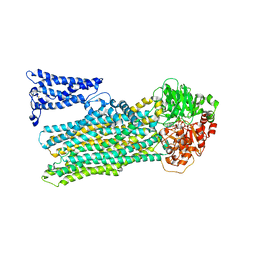 | | Cryo-EM structure of human ABC transporter ABCC2 under active turnover condition | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family C member 2, ... | | Authors: | Mao, Y.X, Chen, Z.P, Wang, L, Hou, W.T, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2023-07-01 | | Release date: | 2024-01-03 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Transport mechanism of human bilirubin transporter ABCC2 tuned by the inter-module regulatory domain.
Nat Commun, 15, 2024
|
|
8JXQ
 
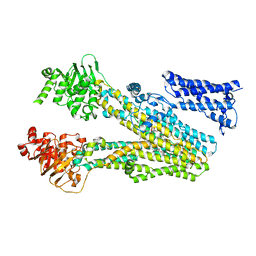 | | Cryo-EM structure of bilirubin ditaurate (BDT) bound human ABC transporter ABCC2 | | Descriptor: | 2-[3-[5-[(E)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-2-[[5-[(3-ethenyl-4-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-3-[3-oxidanylidene-3-(2-sulfoethylamino)propyl]-1H-pyrrol-2-yl]methyl]-4-methyl-1H-pyrrol-3-yl]propanoylamino]ethanesulfonic acid, ATP-binding cassette sub-family C member 2, CHOLESTEROL | | Authors: | Mao, Y.X, Chen, Z.P, Wang, L, Hou, W.T, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2023-07-01 | | Release date: | 2024-01-03 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Transport mechanism of human bilirubin transporter ABCC2 tuned by the inter-module regulatory domain.
Nat Commun, 15, 2024
|
|
3PXG
 
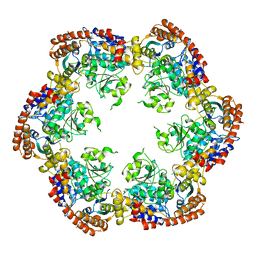 | | Structure of MecA121 and ClpC1-485 complex | | Descriptor: | Adapter protein mecA 1, Negative regulator of genetic competence ClpC/MecB | | Authors: | Wang, F, Mei, Z.Q, Wang, J.W, Shi, Y.G. | | Deposit date: | 2010-12-09 | | Release date: | 2011-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.654 Å) | | Cite: | Structure and mechanism of the hexameric MecA-ClpC molecular machine.
Nature, 471, 2011
|
|
5XAF
 
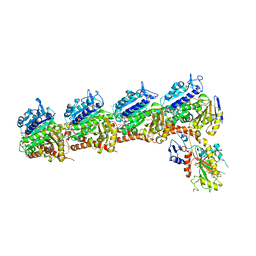 | | Crystal structure of tubulin-stathmin-TTL-Compound Z1 complex | | Descriptor: | (3S,4R)-4-(3-hydroxy-4-methoxyphenyl)-3-methyl-1-(3,4,5-trimethoxyphenyl)azetidin-2-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Zhang, H, Luo, C, Wang, Y. | | Deposit date: | 2017-03-12 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.551 Å) | | Cite: | Design, synthesis, biological evaluation and cocrystal structures with tubulin of chiral beta-lactam bridged combretastatin A-4 analogues as potent antitumor agents
Eur J Med Chem, 144, 2017
|
|
3T7I
 
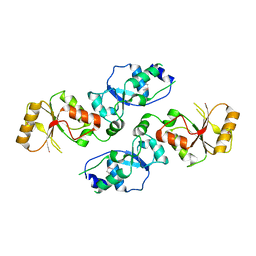 | | Crystal structure of Se-Met Rtt107p (residues 820-1070) | | Descriptor: | Regulator of Ty1 transposition protein 107 | | Authors: | Li, X, Li, F, Wu, J, Shi, Y. | | Deposit date: | 2011-07-30 | | Release date: | 2012-02-15 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of C-terminal Tandem BRCT Repeats of Rtt107 Protein Reveals Critical Role in Interaction with Phosphorylated Histone H2A during DNA Damage Repair
J.Biol.Chem., 287, 2012
|
|
1CYF
 
 | |
6HBX
 
 | |
3JY9
 
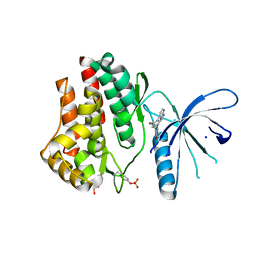 | | Janus Kinase 2 Inhibitors | | Descriptor: | (3S)-3-(4-hydroxyphenyl)-1,5-dihydro-1,5,12-triazabenzo[4,5]cycloocta[1,2,3-cd]inden-4(3H)-one, SODIUM ION, Tyrosine-protein kinase JAK2 | | Authors: | Zuccola, H.J, Ledeboer, M.W, Pierce, A.C. | | Deposit date: | 2009-09-21 | | Release date: | 2009-12-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Janus kinase 2 inhibitors. Synthesis and characterization of a novel polycyclic azaindole.
J.Med.Chem., 52, 2009
|
|
6HC5
 
 | |
7E2R
 
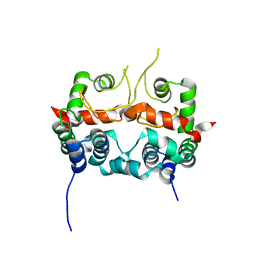 | | The ligand-free structure of Arabidopsis thaliana GUN4 | | Descriptor: | Tetrapyrrole-binding protein, chloroplastic | | Authors: | Liu, L, Hu, J. | | Deposit date: | 2021-02-07 | | Release date: | 2021-08-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of bilin binding by the chlorophyll biosynthesis regulator GUN4.
Protein Sci., 30, 2021
|
|
5WNU
 
 | | Crystal Structure of 30S ribosomal subunit from Thermus thermophilus | | Descriptor: | (1R,2S,3S,4R,6R)-4,6-diamino-2-{[3-O-(2,6-diamino-2,6-dideoxy-alpha-L-altropyranosyl)-beta-L-arabinofuranosyl]oxy}-3-hydroxycyclohexyl 2-amino-2-deoxy-alpha-D-allopyranoside, 16S Ribosomal RNA rRNA, 30S ribosomal protein S10, ... | | Authors: | DeMirci, H. | | Deposit date: | 2017-08-01 | | Release date: | 2018-02-21 | | Last modified: | 2020-10-21 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | 2'-O-methylation in mRNA disrupts tRNA decoding during translation elongation.
Nat. Struct. Mol. Biol., 25, 2018
|
|
5XAG
 
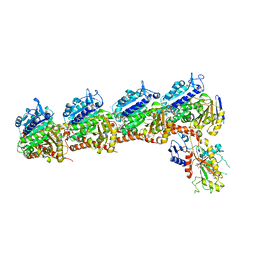 | | Crystal structure of tubulin-stathmin-TTL-Compound Z2 complex | | Descriptor: | (3~{R},4~{R})-3-(hydroxymethyl)-4-(4-methoxy-3-oxidanyl-phenyl)-1-(3,4,5-trimethoxyphenyl)azetidin-2-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Zhang, H, Luo, C, Wang, Y. | | Deposit date: | 2017-03-12 | | Release date: | 2018-01-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Design, synthesis, biological evaluation and cocrystal structures with tubulin of chiral beta-lactam bridged combretastatin A-4 analogues as potent antitumor agents
Eur J Med Chem, 144, 2017
|
|
3QUX
 
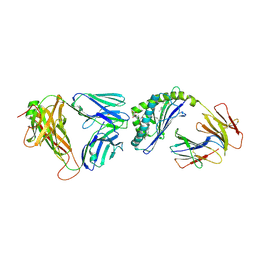 | | Structure of the mouse CD1d-alpha-C-GalCer-iNKT TCR complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Li, Y, Girardi, E, Yu, E.D, Zajonc, D.M. | | Deposit date: | 2011-02-24 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Galactose-modified iNKT cell agonists stabilized by an induced fit of CD1d prevent tumour metastasis.
Embo J., 30, 2011
|
|
3QE7
 
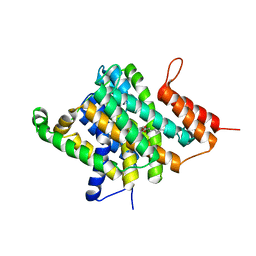 | | Crystal Structure of Uracil Transporter--UraA | | Descriptor: | URACIL, Uracil permease, nonyl beta-D-glucopyranoside | | Authors: | Lu, F.R, Li, S, Yan, N. | | Deposit date: | 2011-01-20 | | Release date: | 2011-03-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.781 Å) | | Cite: | Structure and mechanism of the uracil transporter UraA
Nature, 472, 2011
|
|
6LMS
 
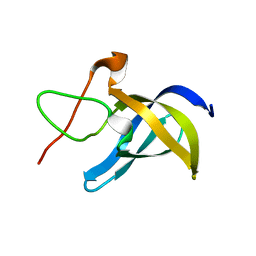 | |
6BOZ
 
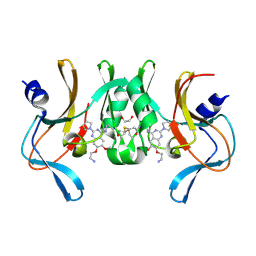 | | Structure of human SETD8 in complex with covalent inhibitor MS4138 | | Descriptor: | 1,2-ETHANEDIOL, N-(3-{[7-(2-aminoethoxy)-6-methoxy-2-(pyrrolidin-1-yl)quinazolin-4-yl]amino}propyl)prop-2-enamide, N-lysine methyltransferase KMT5A | | Authors: | Babault, N, Anqi, M, Jin, J. | | Deposit date: | 2017-11-21 | | Release date: | 2019-05-01 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The dynamic conformational landscape of the protein methyltransferase SETD8.
Elife, 8, 2019
|
|
6LNU
 
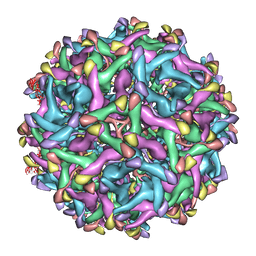 | | Cryo-EM structure of immature Zika virus | | Descriptor: | Genome polyprotein | | Authors: | Tan, T.Y, Fibriansah, G, Kostyuchenko, V.A, Ng, T.S, Lim, X.X, Lim, X.N, Shi, J, Morais, M.C, Corti, D, Lok, S.M. | | Deposit date: | 2020-01-02 | | Release date: | 2020-02-26 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Capsid protein structure in Zika virus reveals the flavivirus assembly process.
Nat Commun, 11, 2020
|
|
5VGW
 
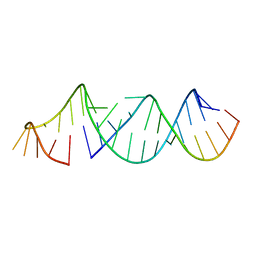 | | RNA hairpin structure containing tetraloop/receptor motif | | Descriptor: | RNA (5'-R(*CP*UP*GP*CP*UP*GP*GP*CP*UP*AP*AP*GP*GP*CP*CP*CP*GP*AP*AP*AP*GP*G)-3'), RNA (5'-R(P*GP*CP*UP*AP*UP*GP*CP*CP*UP*GP*CP*UP*G)-3') | | Authors: | Zhang, W, Szostak, J.W. | | Deposit date: | 2017-04-11 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural Rationale for the Enhanced Catalysis of Nonenzymatic RNA Primer Extension by a Downstream Oligonucleotide.
J. Am. Chem. Soc., 140, 2018
|
|
7E2V
 
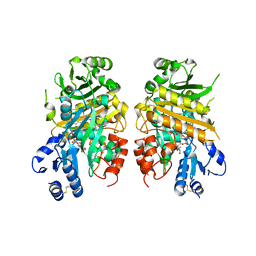 | | Crystal structure of MaDA-3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, MaDA-3 | | Authors: | Gao, L, Du, X, Fan, J, Lei, X. | | Deposit date: | 2021-02-07 | | Release date: | 2021-12-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Enzymatic control of endo- and exo-stereoselective Diels-Alder reactions with broad substrate scope.
Nat Catal, 2021
|
|
2PCB
 
 | |
7DV4
 
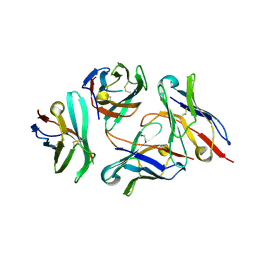 | | Crystal structure of anti-CTLA-4 VH domain in complex with human CTLA-4 | | Descriptor: | 1,2-ETHANEDIOL, 4003-1(VH), Cytotoxic T-lymphocyte protein 4 | | Authors: | Li, H, Gan, X, He, Y. | | Deposit date: | 2021-01-12 | | Release date: | 2022-01-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | An anti-CTLA-4 heavy chain-only antibody with enhanced T reg depletion shows excellent preclinical efficacy and safety profile.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
5Y9V
 
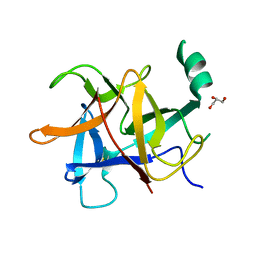 | | Crystal structure of diamondback moth ryanodine receptor N-terminal domain | | Descriptor: | CHLORIDE ION, GLYCEROL, Ryanodine receptor 1 | | Authors: | Lin, L, Yuchi, Z. | | Deposit date: | 2017-08-28 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.841 Å) | | Cite: | Crystal structure of ryanodine receptor N-terminal domain from Plutella xylostella reveals two potential species-specific insecticide-targeting sites.
Insect Biochem. Mol. Biol., 92, 2017
|
|
6LMR
 
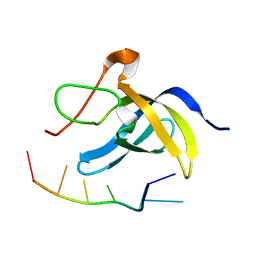 | | Solution structure of cold shock domain and ssDNA complex | | Descriptor: | DNA (5'-D(P*AP*AP*CP*AP*CP*CP*T)-3'), Y-box-binding protein 1 | | Authors: | Fan, J, Yang, D. | | Deposit date: | 2019-12-26 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis of DNA binding to human YB-1 cold shock domain regulated by phosphorylation.
Nucleic Acids Res., 48, 2020
|
|
6LNT
 
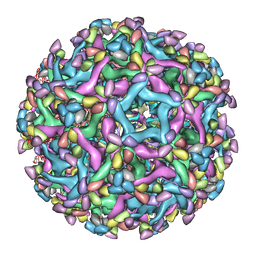 | | Cryo-EM structure of immature Zika virus in complex with human antibody DV62.5 Fab | | Descriptor: | Envelope protein, Fab DV62.5 heavy-chain variable region, Fab DV62.5 light-chain variable region, ... | | Authors: | Tan, T.Y, Fibriansah, G, Kostyuchenko, V.A, Ng, T.S, Lim, X.X, Lim, X.N, Shi, J, Morais, M.C, Corti, D, Lok, S.M. | | Deposit date: | 2020-01-02 | | Release date: | 2020-02-26 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Capsid protein structure in Zika virus reveals the flavivirus assembly process.
Nat Commun, 11, 2020
|
|
