3EKR
 
 | | Dihydroxylphenyl amides as inhibitors of the Hsp90 molecular chaperone | | Descriptor: | 4-{[(2R)-2-(2-methylphenyl)pyrrolidin-1-yl]carbonyl}benzene-1,3-diol, Heat shock protein HSP 90-alpha, PHOSPHATE ION | | Authors: | Gajiwala, K.S. | | Deposit date: | 2008-09-19 | | Release date: | 2008-11-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Dihydroxylphenyl amides as inhibitors of the Hsp90 molecular chaperone.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
7JYM
 
 | | CRYSTAL STRUCTURE OF RAR-RELATED ORPHAN RECEPTOR C (NHIS-RORGT(244-487)-L6-SRC1(678-692)) IN COMPLEX WITH A TRICYCLIC SULFONE INVERSE AGONIST | | Descriptor: | (3R,5S)-3-fluoro-5-[(3aR,9bR)-9b-[(4-fluorophenyl)sulfonyl]-7-(1,1,1,2,3,3,3-heptafluoropropan-2-yl)-1,2,3a,4,5,9b-hexahydro-3H-benzo[e]indole-3-carbonyl]-1-(2-hydroxy-2-methylpropyl)pyrrolidin-2-one, Nuclear receptor ROR-gamma, Nuclear receptor coactivator 1 | | Authors: | Sack, J. | | Deposit date: | 2020-08-31 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.051 Å) | | Cite: | Novel Tricyclic Pyroglutamide Derivatives as Potent ROR gamma t Inverse Agonists Identified using a Virtual Screening Approach.
Acs Med.Chem.Lett., 11, 2020
|
|
7EO0
 
 | | FOOT AND MOUTH DISEASE VIRUS O/TIBET/99-BOUND THE SINGLE CHAIN FRAGMEN ANTIBODY C4 | | Descriptor: | Ig heavy chain variable region, Ig lamda chain variable region, O/TIBET/99 VP1, ... | | Authors: | He, Y, Li, K. | | Deposit date: | 2021-04-21 | | Release date: | 2021-08-18 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Two Cross-Protective Antigen Sites on Foot-and-Mouth Disease Virus Serotype O Structurally Revealed by Broadly Neutralizing Antibodies from Cattle.
J.Virol., 95, 2021
|
|
4IS7
 
 | |
6IYF
 
 | | Structure of pSTING complex | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, SULFATE ION, Stimulator of interferon genes protein | | Authors: | Yuan, Z.L, Shang, G.J, Cong, X.Y, Gu, L.C. | | Deposit date: | 2018-12-15 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.764 Å) | | Cite: | Crystal structures of porcine STINGCBD-CDN complexes reveal the mechanism of ligand recognition and discrimination of STING proteins.
J.Biol.Chem., 294, 2019
|
|
4G6H
 
 | | Crystal structure of NDH with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION, ... | | Authors: | Li, W, Feng, Y, Ge, J, Yang, M. | | Deposit date: | 2012-07-19 | | Release date: | 2012-10-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.262 Å) | | Cite: | Structural insight into the type-II mitochondrial NADH dehydrogenases.
Nature, 491, 2012
|
|
4G6G
 
 | | Crystal structure of NDH with TRT | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, FRAGMENT OF TRITON X-100, MAGNESIUM ION, ... | | Authors: | Li, W, Feng, Y, Ge, J, Yang, M. | | Deposit date: | 2012-07-19 | | Release date: | 2012-10-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural insight into the type-II mitochondrial NADH dehydrogenases.
Nature, 491, 2012
|
|
4G74
 
 | | Crystal structure of NDH with Quinone | | Descriptor: | 2,3-DIMETHOXY-5-METHYL-6-(3,11,15,19-TETRAMETHYL-EICOSA-2,6,10,14,18-PENTAENYL)-[1,4]BENZOQUINONE, FLAVIN-ADENINE DINUCLEOTIDE, FRAGMENT OF TRITON X-100, ... | | Authors: | Li, W, Feng, Y, Ge, J, Yang, M. | | Deposit date: | 2012-07-19 | | Release date: | 2012-10-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structural insight into the type-II mitochondrial NADH dehydrogenases.
Nature, 491, 2012
|
|
6CB1
 
 | | Yeast nucleolar pre-60S ribosomal subunit (state 3) | | Descriptor: | 35S pre-ribosomal RNA miscRNA, 5.8S rRNA, 60S ribosomal protein L13-A, ... | | Authors: | Sanghai, Z.A, Miller, L, Barandun, J, Hunziker, M, Chaker-Margot, M, Klinge, S. | | Deposit date: | 2018-02-01 | | Release date: | 2018-03-14 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Modular assembly of the nucleolar pre-60S ribosomal subunit.
Nature, 556, 2018
|
|
7BP3
 
 | | Cryo-EM structure of the human MCT2 | | Descriptor: | Monocarboxylate transporter 2 | | Authors: | Zhang, B, Jin, Q, Zhang, X, Guo, J, Ye, S. | | Deposit date: | 2020-03-21 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cooperative transport mechanism of human monocarboxylate transporter 2.
Nat Commun, 11, 2020
|
|
6CMX
 
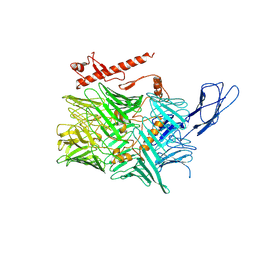 | | Human Teneurin 2 extra-cellular region | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Teneurin-2, alpha-D-mannopyranose, ... | | Authors: | Shalev-Benami, M, Li, J, Sudhof, T, Skiniotis, G, Arac, D. | | Deposit date: | 2018-03-06 | | Release date: | 2018-07-25 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural Basis for Teneurin Function in Circuit-Wiring: A Toxin Motif at the Synapse.
Cell, 173, 2018
|
|
4JLK
 
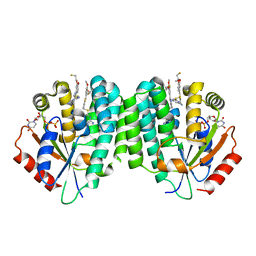 | | Human dCK C4S-S74E mutant in complex with UDP and the F2.2.1 inhibitoR (2-[({2-[3-(2-FLUOROETHOXY)-4-METHOXYPHENYL]-5-METHYL-1,3-THIAZOL-4-YL}METHYL)SULFANYL]PYRIMIDINE-4,6-DIAMINE) | | Descriptor: | 2-[({2-[3-(2-fluoroethoxy)-4-methoxyphenyl]-5-methyl-1,3-thiazol-4-yl}methyl)sulfanyl]pyrimidine-4,6-diamine, Deoxycytidine kinase, URIDINE-5'-DIPHOSPHATE | | Authors: | Nomme, J, Lavie, A. | | Deposit date: | 2013-03-12 | | Release date: | 2013-09-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Development of new deoxycytidine kinase inhibitors and noninvasive in vivo evaluation using positron emission tomography.
J.Med.Chem., 56, 2013
|
|
4JND
 
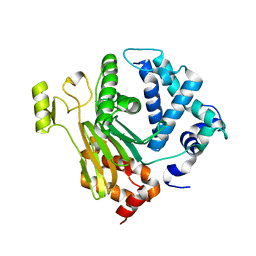 | | Structure of a C.elegans sex determining protein | | Descriptor: | Ca(2+)/calmodulin-dependent protein kinase phosphatase, MAGNESIUM ION | | Authors: | Feng, Y, Zhang, Y, Ge, J, Yang, M. | | Deposit date: | 2013-03-15 | | Release date: | 2013-06-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.652 Å) | | Cite: | Structural insight into Caenorhabditis elegans sex-determining protein FEM-2.
J.Biol.Chem., 288, 2013
|
|
8G8C
 
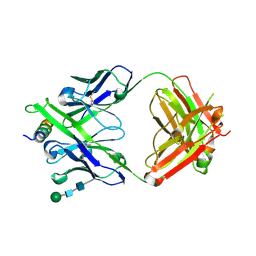 | | Crystal structure of DH1322.1 Fab in complex with HIV proximal MPER peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DH1322.1 heavy chain, DH1322.1 light chain, ... | | Authors: | Niyongabo, A, Janus, B.M, Ofek, G. | | Deposit date: | 2023-02-17 | | Release date: | 2024-05-22 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Vaccine induction of heterologous HIV-1-neutralizing antibody B cell lineages in humans.
Cell, 187, 2024
|
|
8G8A
 
 | | Crystal structure of DH1317.8 Fab in complex with HIV proximal MPER peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DH1317.8 heavy chain, DH1317.8 light chain, ... | | Authors: | Janus, B.M, Astavans, A, Ofek, G. | | Deposit date: | 2023-02-17 | | Release date: | 2024-05-22 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Vaccine induction of heterologous HIV-1-neutralizing antibody B cell lineages in humans.
Cell, 187, 2024
|
|
8G8D
 
 | | Crystal structure of DH1346 Fab in complex with HIV proximal MPER peptide | | Descriptor: | DH1346 heavy chain, DH1346 light chain, FLUORIDE ION, ... | | Authors: | Niyongabo, A, Janus, B.M, Ofek, G. | | Deposit date: | 2023-02-17 | | Release date: | 2024-05-22 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Vaccine induction of heterologous HIV-1-neutralizing antibody B cell lineages in humans.
Cell, 187, 2024
|
|
5Z5K
 
 | | Structure of the DCC-Draxin complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Draxin, ... | | Authors: | Liu, Y, Xiao, J, Wang, J. | | Deposit date: | 2018-01-18 | | Release date: | 2018-06-20 | | Last modified: | 2021-03-10 | | Method: | X-RAY DIFFRACTION (2.493 Å) | | Cite: | Structural Basis for Draxin-Modulated Axon Guidance and Fasciculation by Netrin-1 through DCC.
Neuron, 97, 2018
|
|
5VND
 
 | | Crystal structure of FGFR1-Y563C (FGFR4 surrogate) covalently bound to H3B-6527 | | Descriptor: | 1,2-ETHANEDIOL, Fibroblast growth factor receptor 1, N-{2-[(6-{[(2,6-dichloro-3,5-dimethoxyphenyl)carbamoyl](methyl)amino}pyrimidin-4-yl)amino]-5-(4-ethylpiperazin-1-yl)phenyl}propanamide, ... | | Authors: | Tsai, J.H.C, Reynolds, D, Fekkes, P, Smith, P, Larsen, N.A. | | Deposit date: | 2017-04-30 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | H3B-6527 Is a Potent and Selective Inhibitor of FGFR4 in FGF19-Driven Hepatocellular Carcinoma.
Cancer Res., 77, 2017
|
|
1CPD
 
 | |
4UB0
 
 | |
4L5B
 
 | | Human dCK C4S-S74E mutant in complex with UDP and the DI-43 inhibitor | | Descriptor: | 1-[5-(4-{[(4,6-diaminopyrimidin-2-yl)sulfanyl]methyl}-5-propyl-1,3-thiazol-2-yl)-2-methoxyphenoxy]-2-methylpropan-2-ol, Deoxycytidine kinase, URIDINE-5'-DIPHOSPHATE | | Authors: | Nomme, J, Lavie, A. | | Deposit date: | 2013-06-10 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Development of new deoxycytidine kinase inhibitors and noninvasive in vivo evaluation using positron emission tomography.
J.Med.Chem., 56, 2013
|
|
1CPG
 
 | |
1CPE
 
 | |
5ZQY
 
 | | Crystal structure of a poly(ADP-ribose) glycohydrolase | | Descriptor: | MAGNESIUM ION, Poly(ADP-ribose) glycohydrolase ARH3, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Wang, M, Yuan, Z, Ma, Y, Wang, J, Liu, X. | | Deposit date: | 2018-04-20 | | Release date: | 2018-08-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.577 Å) | | Cite: | Structure-function analyses reveal the mechanism of the ARH3-dependent hydrolysis of ADP-ribosylation.
J. Biol. Chem., 293, 2018
|
|
6C0F
 
 | | Yeast nucleolar pre-60S ribosomal subunit (state 2) | | Descriptor: | 5.8S rRNA, 60S ribosomal protein L13-A, 60S ribosomal protein L14-A, ... | | Authors: | Sanghai, Z.A, Miller, L, Barandun, J, Hunziker, M, Chaker-Margot, M, Klinge, S. | | Deposit date: | 2017-12-29 | | Release date: | 2018-03-14 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Modular assembly of the nucleolar pre-60S ribosomal subunit.
Nature, 556, 2018
|
|
