1N11
 
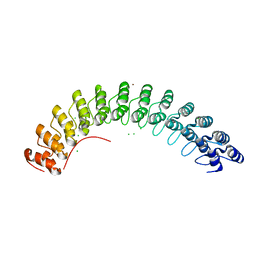 | | D34 REGION OF HUMAN ANKYRIN-R AND LINKER | | Descriptor: | Ankyrin, BROMIDE ION, CHLORIDE ION | | Authors: | Michaely, P, Tomchick, D.R, Machius, M, Anderson, R.G.W. | | Deposit date: | 2002-10-16 | | Release date: | 2002-12-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a 12 ANK repeat stack from human ankyrinR
Embo J., 21, 2002
|
|
1OLU
 
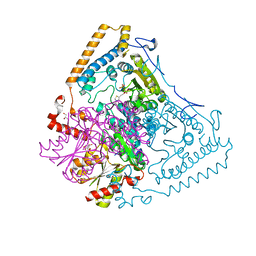 | | Roles of His291-alpha and His146-beta' in the reductive acylation reaction catalyzed by human branched-chain alpha-ketoacid dehydrogenase | | Descriptor: | 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA SUBUNIT, GLYCEROL, ... | | Authors: | Wynn, R.M, Machius, M, Chuang, J.L, Li, J, Tomchick, D.R, Chuang, D.T. | | Deposit date: | 2003-08-15 | | Release date: | 2003-08-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Roles of His291-Alpha and His146-Beta' in the Reductive Acylation Reaction Catalyzed by Human Branched-Chain Alpha-Ketoacid Dehydrogenase: Refined Phosphorylation Loop Structure in the Active Site.
J.Biol.Chem., 278, 2003
|
|
1OLX
 
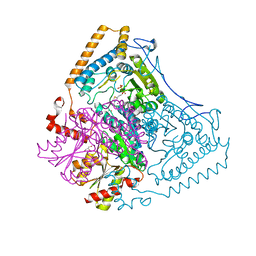 | | Roles of His291-alpha and His146-beta' in the reductive acylation reaction catalyzed by human branched-chain alpha-ketoacid dehydrogenase | | Descriptor: | 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA SUBUNIT, GLYCEROL, ... | | Authors: | Wynn, R.M, Machius, M, Chuang, J.L, Li, J, Tomchick, D.R, Chuang, D.T. | | Deposit date: | 2003-08-18 | | Release date: | 2003-08-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Roles of His291-Alpha and His146-Beta' in the Reductive Acylation Reaction Catalyzed by Human Branched-Chain Alpha-Ketoacid Dehydrogenase: Refined Phosphorylation Loop Structure in the Active Site.
J.Biol.Chem., 278, 2003
|
|
1OLS
 
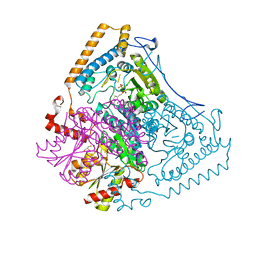 | | Roles of His291-alpha and His146-beta' in the reductive acylation reaction catalyzed by human branched-chain alpha-ketoacid dehydrogenase | | Descriptor: | 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA SUBUNIT, GLYCEROL, ... | | Authors: | Wynn, R.M, Machius, M, Chuang, J.L, Li, J, Tomchick, D.R, Chuang, D.T. | | Deposit date: | 2003-08-12 | | Release date: | 2003-08-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Roles of His291-Alpha and His146-Beta' in the Reductive Acylation Reaction Catalyzed by Human Branched-Chain Alpha-Ketoacid Dehydrogenase: Refined Phosphorylation Loop Structure in the Active Site.
J.Biol.Chem., 278, 2003
|
|
2IW5
 
 | | Structural Basis for CoREST-Dependent Demethylation of Nucleosomes by the Human LSD1 Histone Demethylase | | Descriptor: | AMMONIUM ION, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Yang, M, Gocke, C.B, Luo, X, Borek, D, Tomchick, D.R, Machius, M, Otwinowski, Z, Yu, H. | | Deposit date: | 2006-06-26 | | Release date: | 2006-08-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structural Basis for Corest-Dependent Demethylation of Nucleosomes by the Human Lsd1 Histone Demethylase
Mol.Cell, 23, 2006
|
|
5TVM
 
 | | Crystal structure of Trypanosoma brucei AdoMetDC/prozyme heterodimer | | Descriptor: | 1,4-DIAMINOBUTANE, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, S-adenosylmethionine decarboxylase alpha chain, ... | | Authors: | Volkov, O.A, Chen, Z, Tomchick, D.R, Phillips, M.A. | | Deposit date: | 2016-11-09 | | Release date: | 2017-01-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.408 Å) | | Cite: | Relief of autoinhibition by conformational switch explains enzyme activation by a catalytically dead paralog.
Elife, 5, 2016
|
|
5U2P
 
 | | The crystal structure of Tp0737 from Treponema pallidum | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Brautigam, C.A, Deka, R.K, Tomchick, D.R, Norgard, M.V. | | Deposit date: | 2016-11-30 | | Release date: | 2017-02-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Functional clues from the crystal structure of an orphan periplasmic ligand-binding protein from Treponema pallidum.
Protein Sci., 26, 2017
|
|
5TVF
 
 | | Crystal structure of Trypanosoma brucei AdoMetDC/prozyme heterodimer in complex with inhibitor CGP 40215 | | Descriptor: | 1,4-DIAMINOBUTANE, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-[C-[N'-(3-CARBAMIMIDOYL-BENZYLIDENIUM)-HYDRAZINO]-[[AMINOMETHYLIDENE]AMINIUM]-IMINOMETHYL]-BENZAMIDINIUM, ... | | Authors: | Phillips, M.A, Volkov, O.A, Chen, Z, Tomchick, D.R. | | Deposit date: | 2016-11-08 | | Release date: | 2017-01-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Relief of autoinhibition by conformational switch explains enzyme activation by a catalytically dead paralog.
Elife, 5, 2016
|
|
3PJN
 
 | | The crystal structure of Tp34 bound to Zn(II) ion at pH 7.5 | | Descriptor: | 1,2-ETHANEDIOL, 34 kDa membrane antigen, SULFATE ION, ... | | Authors: | Brautigam, C.A, Deka, R.K, Tomchick, D.R, Machius, M, Norgard, M.V. | | Deposit date: | 2010-11-10 | | Release date: | 2011-11-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Characterization of the Tp34 lipoprotein from Treponema pallidum suggests a role in transition metal homeostasis
To be Published
|
|
3PJL
 
 | | The crystal structure of Tp34 bound to Co (II) ion at pH 7.5 | | Descriptor: | 1,2-ETHANEDIOL, 34 kDa membrane antigen, COBALT (II) ION, ... | | Authors: | Brautigam, C.A, Deka, R.K, Tomchick, D.R, Machius, M, Norgard, M.V. | | Deposit date: | 2010-11-10 | | Release date: | 2011-11-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Characterization of the Tp34 lipoprotein from Treponema pallidum suggests a role in transition metal homeostasis
To be Published
|
|
3LET
 
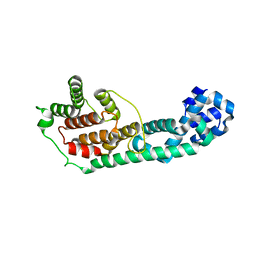 | | Crystal Structure of Fic domain containing AMPylator, VopS | | Descriptor: | Adenosine monophosphate-protein transferase vopS | | Authors: | Luong, P.H, Kinch, L.N, Brautigam, C.A, Grishin, N.V, Tomchick, D.R, Orth, K. | | Deposit date: | 2010-01-15 | | Release date: | 2010-04-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Structural and Kinetic Analysis of VopS with Fic Domain Supports a Direct Transfer Mechanism for AMPylation
To be Published
|
|
3N29
 
 | | Crystal structure of carboxynorspermidine decarboxylase complexed with Norspermidine from Campylobacter jejuni | | Descriptor: | Carboxynorspermidine decarboxylase, GLYCEROL, N-(3-aminopropyl)propane-1,3-diamine, ... | | Authors: | Deng, X, Lee, J, Michael, A.J, Tomchick, D.R, Goldsmith, E.J, Phillips, M.A. | | Deposit date: | 2010-05-17 | | Release date: | 2010-06-09 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Evolution of substrate specificity within a diverse family of beta/alpha-barrel-fold basic amino acid decarboxylases: X-ray structure determination of enzymes with specificity for L-arginine and carboxynorspermidine.
J.Biol.Chem., 285, 2010
|
|
3N2O
 
 | | X-ray crystal structure of arginine decarboxylase complexed with Arginine from Vibrio vulnificus | | Descriptor: | AGMATINE, Biosynthetic arginine decarboxylase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Deng, X, Lee, J, Michael, A.J, Tomchick, D.R, Goldsmith, E.J, Phillips, M.A. | | Deposit date: | 2010-05-18 | | Release date: | 2010-06-09 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Evolution of substrate specificity within a diverse family of beta/alpha-barrel-fold basic amino acid decarboxylases: X-ray structure determination of enzymes with specificity for L-arginine and carboxynorspermidine.
J.Biol.Chem., 285, 2010
|
|
6MRW
 
 | | 14-meric ClyA pore complex | | Descriptor: | Hemolysin E, chromosomal | | Authors: | Peng, W, de Souza Santos, M, Li, Y, Tomchick, D.R, Orth, K. | | Deposit date: | 2018-10-15 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | High-resolution cryo-EM structures of the E. coli hemolysin ClyA oligomers.
Plos One, 14, 2019
|
|
6MRU
 
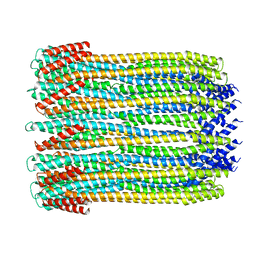 | | 13-meric ClyA pore complex | | Descriptor: | Hemolysin E, chromosomal | | Authors: | Peng, W, de Souza Santos, M, Li, Y, Tomchick, D.R, Orth, K. | | Deposit date: | 2018-10-15 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | High-resolution cryo-EM structures of the E. coli hemolysin ClyA oligomers.
Plos One, 14, 2019
|
|
6MRT
 
 | | 12-meric ClyA pore complex | | Descriptor: | Hemolysin E, chromosomal | | Authors: | Peng, W, de Souza Santos, M, Li, Y, Tomchick, D.R, Orth, K. | | Deposit date: | 2018-10-15 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | High-resolution cryo-EM structures of the E. coli hemolysin ClyA oligomers.
Plos One, 14, 2019
|
|
1GKZ
 
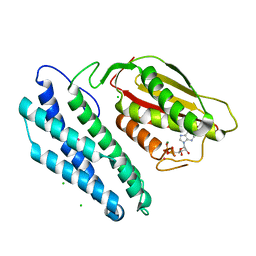 | | Branched-chain alpha-ketoacid dehydrogenase kinase (BCK) complxed with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Machius, M, Chuang, J.L, Wynn, R.M, Tomchick, D.R, Chuang, D.T. | | Deposit date: | 2001-08-22 | | Release date: | 2001-10-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Rat Bckd Kinase: Nucleotide-Induced Domain Communication in a Mitochondrial Protein Kinase.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1FHG
 
 | | HIGH RESOLUTION REFINEMENT OF TELOKIN | | Descriptor: | TELOKIN | | Authors: | Tomchick, D.R, Minor, W, Kiyatkin, A, Lewinski, K, Somlyo, A.V, Somlyo, A.P. | | Deposit date: | 2000-08-01 | | Release date: | 2000-08-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure determination of telokin, the C-terminal domain of myosin light chain kinase, at 2.8 A resolution.
J.Mol.Biol., 227, 1992
|
|
7UT5
 
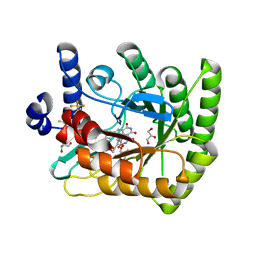 | | Acinetobacter baumannii dihydroorotate dehydrogenase bound with inhibitor DSM186 | | Descriptor: | (4R)-7-methyl-N-[4-(pentafluoro-lambda~6~-sulfanyl)phenyl]imidazo[1,2-a]pyrimidin-5-amine, Dihydroorotate dehydrogenase (quinone), FLAVIN MONONUCLEOTIDE, ... | | Authors: | Deng, X, Phillips, M, Tomchick, D. | | Deposit date: | 2022-04-26 | | Release date: | 2022-12-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Repurposed dihydroorotate dehydrogenase inhibitors with efficacy against drug-resistant Acinetobacter baumannii.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
5BOO
 
 | | Crystal structure of Plasmodium falciparum dihydroorotate dehydrogenase bound with Inhibitor DSM265 | | Descriptor: | 2-(1,1-difluoroethyl)-5-methyl-N-[4-(pentafluoro-lambda~6~-sulfanyl)phenyl][1,2,4]triazolo[1,5-a]pyrimidin-7-amine, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Phillips, M, Deng, X, Tomchick, D. | | Deposit date: | 2015-05-27 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A long-duration dihydroorotate dehydrogenase inhibitor (DSM265) for prevention and treatment of malaria.
Sci Transl Med, 7, 2015
|
|
5FI8
 
 | | Crystal structure of plasmodium falciparum dihydroorotate dehydrogenase bounded with DSM422 (Tetrahydro-2-naphthyl and 2-indanyl triazolopyrimidine) | | Descriptor: | 2-[1,1-bis(fluoranyl)ethyl]-~{N}-[(2~{S})-7-bromanyl-1,2,3,4-tetrahydronaphthalen-2-yl]-5-methyl-[1,2,4]triazolo[1,5-a]pyrimidin-7-amine, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Deng, X, Kokkonda, S, Tomchick, D, Phillips, M. | | Deposit date: | 2015-12-22 | | Release date: | 2016-05-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Tetrahydro-2-naphthyl and 2-Indanyl Triazolopyrimidines Targeting Plasmodium falciparum Dihydroorotate Dehydrogenase Display Potent and Selective Antimalarial Activity.
J.Med.Chem., 59, 2016
|
|
1KIL
 
 | | Three-dimensional structure of the complexin/SNARE complex | | Descriptor: | Complexin I SNARE-complex binding region, MAGNESIUM ION, SNAP-25 C-terminal SNARE motif, ... | | Authors: | Chen, X, Tomchick, D, Kovrigin, E, Arac, D, Machius, M, Sudhof, T.C, Rizo, J. | | Deposit date: | 2001-12-03 | | Release date: | 2002-03-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Three-dimensional structure of the complexin/SNARE complex.
Neuron, 33, 2002
|
|
4RX0
 
 | | Crystal structure of Plasmodium falciparum dihydroorotate dehydrogenase bound with Inhibitor DSM265 | | Descriptor: | 2-(1,1-difluoroethyl)-5-methyl-N-[4-(pentafluoro-lambda~6~-sulfanyl)phenyl][1,2,4]triazolo[1,5-a]pyrimidin-7-amine, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Deng, X, Phillips, M, Tomchick, D. | | Deposit date: | 2014-12-08 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A long-duration dihydroorotate dehydrogenase inhibitor (DSM265) for prevention and treatment of malaria.
Sci Transl Med, 7, 2015
|
|
9DI6
 
 | | Crystal structure of Plasmodium falciparum dihydroorotate dehydrogenase bound with Inhibitor DSM679 (ethyl 1,4-dimethyl-5-((6-(trifluoromethyl)pyridin-3-yl)methyl)-1H-pyrazole-3-carboxylate) | | Descriptor: | 6-[bis(oxidanyl)methyl]-5~{H}-pyrimidine-2,4-dione, CITRIC ACID, Dihydroorotate dehydrogenase (quinone), ... | | Authors: | Deng, X, Tomchick, D, Phillips, M. | | Deposit date: | 2024-09-05 | | Release date: | 2025-01-01 | | Last modified: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structure-Based Discovery and Development of Highly Potent Dihydroorotate Dehydrogenase Inhibitors for Malaria Chemoprevention.
J.Med.Chem., 68, 2025
|
|
9DIK
 
 | | Crystal structure of Plasmodium falciparum dihydroorotate dehydrogenase bound with Inhibitor DSM681 (N-cyclopropyl-1,4-dimethyl-5-((6-(trifluoromethyl)pyridin-3-yl)methyl)-1H-pyrazole-3-carboxamide) | | Descriptor: | 6-[bis(oxidanyl)methyl]-5~{H}-pyrimidine-2,4-dione, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Deng, X, Tomchick, D, Phillips, M. | | Deposit date: | 2024-09-05 | | Release date: | 2025-01-01 | | Last modified: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Discovery and Development of Highly Potent Dihydroorotate Dehydrogenase Inhibitors for Malaria Chemoprevention.
J.Med.Chem., 68, 2025
|
|
