2Z6M
 
 | |
2ZAV
 
 | |
7KFF
 
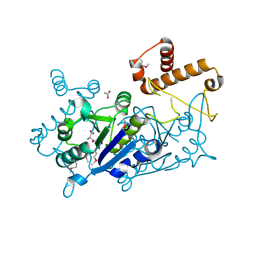 | | Crystal structure of TrmD tRNA (guanine-N1)-methyltransferase from Corynebacterium diphtheriae in complex with SAH | | Descriptor: | ACETATE ION, S-ADENOSYL-L-HOMOCYSTEINE, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Michalska, K, Tanase, L, Maltseva, N, Kim, Y, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-10-13 | | Release date: | 2020-10-28 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of TrmD tRNA (guanine-N1)-methyltransferase from Corynebacterium diphtheriae in complex with SAH
To Be Published
|
|
3WRS
 
 | |
3WRQ
 
 | |
3BUV
 
 | | Crystal structure of human Delta(4)-3-ketosteroid 5-beta-reductase in complex with NADP and HEPES. Resolution: 1.35 A. | | Descriptor: | 3-oxo-5-beta-steroid 4-dehydrogenase, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Di Costanzo, L, Drury, J, Penning, T.M, Christianson, D.W. | | Deposit date: | 2008-01-03 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal Structure of Human Liver {Delta}4-3-Ketosteroid 5{beta}-Reductase (AKR1D1) and Implications for Substrate Binding and Catalysis.
J.Biol.Chem., 283, 2008
|
|
4RDP
 
 | | Crystal structure of Cmr4 | | Descriptor: | CRISPR system Cmr subunit Cmr4 | | Authors: | Shao, Y, Tang, L, Li, H. | | Deposit date: | 2014-09-19 | | Release date: | 2014-12-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Essential Structural and Functional Roles of the Cmr4 Subunit in RNA Cleavage by the Cmr CRISPR-Cas Complex.
Cell Rep, 9, 2014
|
|
3BSQ
 
 | | Crystal structure of human kallikrein 7 produced as a secretion protein in E.coli | | Descriptor: | Kallikrein-7, SULFATE ION | | Authors: | Fernandez, I.S, Standker, L, Magert, H.J, Forssmann, W.G, Gimenez-Gallego, G, Romero, A. | | Deposit date: | 2007-12-26 | | Release date: | 2008-04-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of human epidermal kallikrein 7 (hK7) synthesized directly in its native state in E. coli: insights into the atomic basis of its inhibition by LEKTI domain 6 (LD6)
J.Mol.Biol., 377, 2008
|
|
4RVP
 
 | | Crystal structure of superoxide dismutase from sedum alfredii | | Descriptor: | ZINC ION, superoxide dismutase | | Authors: | Qiu, R, Li, C, Zhai, J, Tang, L, Zhang, H, Yuan, M, Hu, X. | | Deposit date: | 2014-11-27 | | Release date: | 2015-12-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The positive effects of Cd and Cd-Zn relationship in the
Zn-related physiological processes involved in growth in the Zn/Cd hyperaccumulator Sedum alfredii
To be Published
|
|
3BUR
 
 | |
3BV7
 
 | | Crystal structure of Delta(4)-3-ketosteroid 5-beta-reductase in complex with NADP and glycerol. Resolution: 1.79 A. | | Descriptor: | 3-oxo-5-beta-steroid 4-dehydrogenase, GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Di Costanzo, L, Drury, J, Penning, T.M, Christianson, D.W. | | Deposit date: | 2008-01-04 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal Structure of Human Liver {Delta}4-3-Ketosteroid 5{beta}-Reductase (AKR1D1) and Implications for Substrate Binding and Catalysis.
J.Biol.Chem., 283, 2008
|
|
3CYU
 
 | | Human Carbonic Anhydrase II complexed with Cryptophane biosensor and xenon | | Descriptor: | Carbonic anhydrase 2, MoMo-2-[4-(2-(4-(methoxy)-1H-1,2,3-triazol-1-yl)ethyl)benzenesulfonamide]-7,12-bis-[3-(4-(methoxy)-1H-1,2,3-triazol-1-y l)propanoic acid]-cryptophane-A, PoPo-2-[4-(2-(4-(methoxy)-1H-1,2,3-triazol-1-yl)ethyl)benzenesulfonamide]-7,12-bis-[3-(4-(methoxy)-1H-1,2,3-triazol-1-yl)propanoic acid]-cryptophane-A, ... | | Authors: | Aaron, J.A, Jude, K.M, Di Costanzo, L, Christianson, D.W. | | Deposit date: | 2008-04-26 | | Release date: | 2008-05-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of a 129Xe-cryptophane biosensor complexed with human carbonic anhydrase II.
J.Am.Chem.Soc., 130, 2008
|
|
3WRO
 
 | |
3WRN
 
 | |
3COT
 
 | | Crystal structure of human liver delta(4)-3-ketosteroid 5beta-reductase (akr1d1) in complex with progesterone and nadp. Resolution: 2.03 A. | | Descriptor: | 3-oxo-5-beta-steroid 4-dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROGESTERONE | | Authors: | Di Costanzo, L, Drury, J, Penning, T.M, Christianson, D.W. | | Deposit date: | 2008-03-29 | | Release date: | 2008-04-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal Structure of Human Liver {Delta}4-3-Ketosteroid 5{beta}-Reductase (AKR1D1) and Implications for Substrate Binding and Catalysis.
J.Biol.Chem., 283, 2008
|
|
3WRR
 
 | |
3ZN2
 
 | | protein engineering of halohydrin dehalogenase | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, ACETATE ION, HALOHYDRIN DEHALOGENASE, ... | | Authors: | Schallmey, M, Jekel, P, Tang, L, Majeric-Elenkov, M, Hoeffken, H.W, Hauer, B, Janssen, D.B. | | Deposit date: | 2013-02-13 | | Release date: | 2014-03-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Single Point Mutation Enhances Hydroxynitrile Synthesis by Halohydrin Dehalogenase.
Enzyme.Microb.Technol., 70, 2015
|
|
5J7S
 
 | | Crystal structure of SM1-71 bound to TAK1-TAB1 | | Descriptor: | Mitogen-activated protein kinase kinase kinase 7/TGF-beta-activated kinase 1 and MAP3K7-binding protein 1 chimera, N-{2-[(5-chloro-2-{[4-(4-methylpiperazin-1-yl)phenyl]amino}pyrimidin-4-yl)amino]phenyl}propanamide | | Authors: | Gurbani, D, Westover, K.D. | | Deposit date: | 2016-04-06 | | Release date: | 2017-02-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.368 Å) | | Cite: | Structure-guided development of covalent TAK1 inhibitors.
Bioorg. Med. Chem., 25, 2017
|
|
5JH6
 
 | | Crystal structure of TL10-92 bound to TAK1-TAB1 | | Descriptor: | 2-[(5-chloro-2-{[4-(4-methylpiperazin-1-yl)phenyl]amino}pyrimidin-4-yl)amino]phenyl prop-2-enoate, Mitogen-activated protein kinase kinase kinase 7,TGF-beta-activated kinase 1 and MAP3K7-binding protein 1 | | Authors: | Gurbani, D, Westover, K.D. | | Deposit date: | 2016-04-20 | | Release date: | 2017-02-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.365 Å) | | Cite: | Structure-guided development of covalent TAK1 inhibitors.
Bioorg. Med. Chem., 25, 2017
|
|
5JK3
 
 | | Crystal structure of TL11-128 bound to TAK1-TAB1 | | Descriptor: | Mitogen-activated protein kinase kinase kinase 7,TGF-beta-activated kinase 1 and MAP3K7-binding protein 1, ~{N}-[2-[5-chloranyl-2-[(1-methylpyrazol-4-yl)amino]pyrimidin-4-yl]oxyphenyl]prop-2-enamide | | Authors: | Gurbani, D, Westover, K.D. | | Deposit date: | 2016-04-25 | | Release date: | 2017-02-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.371 Å) | | Cite: | Structure-guided development of covalent TAK1 inhibitors.
Bioorg. Med. Chem., 25, 2017
|
|
5J8I
 
 | | Crystal structure of TL11-113 bound to TAK1-TAB1 | | Descriptor: | Mitogen-activated protein kinase kinase kinase 7/TGF-beta-activated kinase 1 and MAP3K7-binding protein 1 chimera, N-{2-[(5-chloro-2-{[4-(4-methylpiperazin-1-yl)phenyl]amino}pyrimidin-4-yl)oxy]phenyl}prop-2-enamide | | Authors: | Gurbani, D, Westover, K.D. | | Deposit date: | 2016-04-07 | | Release date: | 2017-02-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.404 Å) | | Cite: | Structure-guided development of covalent TAK1 inhibitors.
Bioorg. Med. Chem., 25, 2017
|
|
5J9L
 
 | | Crystal structure of CPT1691 bound to TAK1-TAB1 | | Descriptor: | Mitogen-activated protein kinase kinase kinase 7,TGF-beta-activated kinase 1 and MAP3K7-binding protein 1, N-(4-((2-((4-(4-methylpiperazin-1-yl)phenyl)amino)-7H-pyrrolo[2,3-d]pyrimidin-4-yl)oxy)phenyl)acrylamide | | Authors: | Gurbani, D, Westover, K.D. | | Deposit date: | 2016-04-10 | | Release date: | 2017-02-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7515 Å) | | Cite: | Structure-guided development of covalent TAK1 inhibitors.
Bioorg. Med. Chem., 25, 2017
|
|
4QQC
 
 | | Crystal Structure of FGF Receptor (FGFR) 4 Kinase Domain in Complex with FIIN-2, an Irreversible Tyrosine Kinase Inhibitor Capable of Overcoming FGFR Kinase Gate-Keeper Mutations | | Descriptor: | Fibroblast growth factor receptor 4, N-(4-{[3-(3,5-dimethoxyphenyl)-7-{[4-(4-methylpiperazin-1-yl)phenyl]amino}-2-oxo-3,4-dihydropyrimido[4,5-d]pyrimidin-1(2H)-yl]methyl}phenyl)propanamide, SULFATE ION | | Authors: | Huang, Z, Mohammadi, M. | | Deposit date: | 2014-06-27 | | Release date: | 2014-10-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Development of covalent inhibitors that can overcome resistance to first-generation FGFR kinase inhibitors.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4R6V
 
 | | Crystal Structure of FGF Receptor (FGFR) 4 Kinase Harboring the V550L Gate-Keeper Mutation in Complex with FIIN-3, an Irreversible Tyrosine Kinase Inhibitor Capable of Overcoming FGFR kinase Gate-Keeper Mutations | | Descriptor: | Fibroblast growth factor receptor 4, N-[4-({[(2,6-dichloro-3,5-dimethoxyphenyl)carbamoyl](6-{[4-(4-methylpiperazin-1-yl)phenyl]amino}pyrimidin-4-yl)amino}methyl)phenyl]propanamide, SULFATE ION | | Authors: | Huang, Z, Mohammadi, M. | | Deposit date: | 2014-08-26 | | Release date: | 2014-10-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.353 Å) | | Cite: | Development of covalent inhibitors that can overcome resistance to first-generation FGFR kinase inhibitors.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4R5S
 
 | | Crystal structure of EGFR 696-1022 L858R in complex with FIIN-3 | | Descriptor: | Epidermal growth factor receptor, N-[4-({[(2,6-dichloro-3,5-dimethoxyphenyl)carbamoyl](6-{[4-(4-methylpiperazin-1-yl)phenyl]amino}pyrimidin-4-yl)amino}methyl)phenyl]propanamide | | Authors: | Zhu, S.J, Yun, C.H. | | Deposit date: | 2014-08-22 | | Release date: | 2014-11-12 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | Development of covalent inhibitors that can overcome resistance to first-generation FGFR kinase inhibitors.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
