2ZAX
 
 | | Crystal Structure of Ferric Cytochrome P450cam | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CAMPHOR, Cytochrome P450-cam, ... | | Authors: | Sakurai, K, Shimada, H, Harada, K, Hayashi, T, Tsukihara, T. | | Deposit date: | 2007-10-11 | | Release date: | 2008-01-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Evaluation of the functional role of the heme-6-propionate side chain in cytochrome P450cam
J.Am.Chem.Soc., 130, 2008
|
|
2Z97
 
 | | Crystal Structure of Ferric Cytochrome P450cam Reconstituted with 7-Methyl-7-depropionated Hemin | | Descriptor: | 7-METHYL-7-DEPROPIONATEHEMIN, CAMPHOR, Cytochrome P450-cam, ... | | Authors: | Hayashi, T, Harada, K, Sakurai, K, Hirota, S, Shimada, H. | | Deposit date: | 2007-09-18 | | Release date: | 2008-09-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A role of the heme-7-propionate side chain in cytochrome P450cam as a gate for regulating the access of water molecules to the substrate-binding site
J.Am.Chem.Soc., 131, 2009
|
|
2ZWU
 
 | | Crystal Structure of Camphor Soaked Ferric Cytochrome P450cam | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CAMPHOR, Camphor 5-monooxygenase, ... | | Authors: | Sakurai, K, Shimada, H, Hayashi, T, Tsukihara, T. | | Deposit date: | 2008-12-18 | | Release date: | 2009-02-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Substrate binding induces structural changes in cytochrome P450cam
Acta Crystallogr.,Sect.F, 65, 2009
|
|
2ZWT
 
 | | Crystal Structure of Ferric Cytochrome P450cam | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CAMPHOR, Camphor 5-monooxygenase, ... | | Authors: | Sakurai, K, Shimada, H, Hayashi, T, Tsukihara, T. | | Deposit date: | 2008-12-18 | | Release date: | 2009-02-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Substrate binding induces structural changes in cytochrome P450cam
Acta Crystallogr.,Sect.F, 65, 2009
|
|
3A25
 
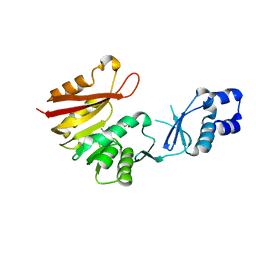 | | Crystal structure of P. horikoshii TYW2 in complex with AdoMet | | Descriptor: | S-ADENOSYLMETHIONINE, Uncharacterized protein PH0793 | | Authors: | Umitsu, M, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2009-04-28 | | Release date: | 2009-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of AdoMet-dependent aminocarboxypropyl transfer reaction catalyzed by tRNA-wybutosine synthesizing enzyme, TYW2
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3VPZ
 
 | |
8JF5
 
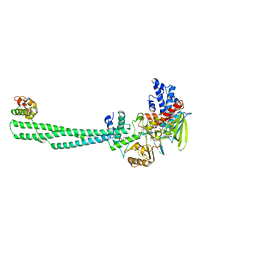 | | Crystal structure of Lysine Specific Demethylase 1 (LSD1) with TAS1440 | | Descriptor: | 4-[5-[(3~{R})-3-azanylpyrrolidin-1-yl]carbonyl-2-[2-fluoranyl-4-(2-methyl-2-oxidanyl-propyl)phenyl]phenyl]-2-fluoranyl-benzenecarbonitrile, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Fukushima, H, Machida, T, Yamashita, S, Suzuki, T. | | Deposit date: | 2023-05-17 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | TAS1440, a histone H3 competitive LSD1 inhibitor, activates both TGF-beta and notch signaling pathways via INSM1 dissociation in neuroendocrine small cell lung cancer
To Be Published
|
|
8BQU
 
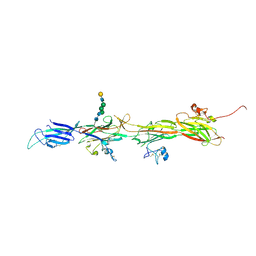 | | Molecular basis of ZP3/ZP1 heteropolymerization: crystal structure of a native vertebrate egg coat filament | | Descriptor: | Choriogenin H, Zona pellucida sperm-binding protein 3, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Bokhove, M, de Sanctis, D, Yasumasu, S, Jovine, L. | | Deposit date: | 2022-11-21 | | Release date: | 2024-03-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | ZP2 cleavage blocks polyspermy by modulating the architecture of the egg coat.
Cell, 187, 2024
|
|
2P6L
 
 | | Crystal structure of PH0725 from Pyrococcus horikoshii OT3 | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, diphthine synthase | | Authors: | Yamamoto, H, Matsuura, Y, Ono, N, Shimada, H, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-19 | | Release date: | 2007-09-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of PH0725 from Pyrococcus horikoshii OT3
To be Published
|
|
3L3F
 
 | |
1GE9
 
 | | SOLUTION STRUCTURE OF THE RIBOSOME RECYCLING FACTOR | | Descriptor: | RIBOSOME RECYCLING FACTOR | | Authors: | Yoshida, T, Uchiyama, S, Nakano, H, Kashimori, H, Kijima, H, Ohshima, T, Saihara, Y, Ishino, T, Shimahara, T, Yoshida, T, Yokose, K, Ohkubo, T, Kaji, A, Kobayashi, Y. | | Deposit date: | 2000-10-19 | | Release date: | 2001-05-16 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the ribosome recycling factor from Aquifex aeolicus.
Biochemistry, 40, 2001
|
|
3AML
 
 | | Structure of the Starch Branching Enzyme I (BEI) from Oryza sativa L | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, BETA-MERCAPTOETHANOL, ... | | Authors: | Kakuta, Y, Chaen, K, Noguchi, J, Vu, N, Kimura, M. | | Deposit date: | 2010-08-20 | | Release date: | 2011-09-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the branching enzyme I (BEI) from Oryza sativa L with implications for catalysis and substrate binding.
Glycobiology, 21, 2011
|
|
3AMK
 
 | | Structure of the Starch Branching Enzyme I (BEI) from Oryza sativa L | | Descriptor: | GLYCEROL, Os06g0726400 protein, PHOSPHATE ION | | Authors: | Kakuta, Y, Chaen, K, Noguchi, J, Vu, N, Kimura, M. | | Deposit date: | 2010-08-20 | | Release date: | 2011-09-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the branching enzyme I (BEI) from Oryza sativa L with implications for catalysis and substrate binding.
Glycobiology, 21, 2011
|
|
8W85
 
 | | HLA-DQ2.5-gamma2 gliadin peptide in complex with DQN0385AE01 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DQN0385AE01 Fab heavy chain, DQN0385AE01 Fab light chain, ... | | Authors: | Irie, M, Tsushima, T, Teranishi-Ikawa, Y, Takahashi, N, Ishii, S, Okura, Y, Fukami, T.A, Torizawa, T. | | Deposit date: | 2023-08-31 | | Release date: | 2023-11-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.769 Å) | | Cite: | Characterizations of a neutralizing antibody broadly reactive to multiple gluten peptide:HLA-DQ2.5 complexes in the context of celiac disease.
Nat Commun, 14, 2023
|
|
8W83
 
 | | HLA-DQ2.5-alpha1 gliadin peptide in complex with DQN0344AE02 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DQN0344AE02 Fab heavy chain, DQN0344AE02 Fab light chain, ... | | Authors: | Irie, M, Tsushima, T, Teranishi-Ikawa, Y, Takahashi, N, Ishii, S, Okura, Y, Fukami, T.A, Torizawa, T. | | Deposit date: | 2023-08-31 | | Release date: | 2023-11-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.818 Å) | | Cite: | Characterizations of a neutralizing antibody broadly reactive to multiple gluten peptide:HLA-DQ2.5 complexes in the context of celiac disease.
Nat Commun, 14, 2023
|
|
8W86
 
 | | HLA-DQ2.5-B/C hordein peptide in complex with DQN0385AE02 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DQN0385AE02 Fab heavy chain, DQN0385AE02 Fab light chain, ... | | Authors: | Irie, M, Tsushima, T, Teranishi-Ikawa, Y, Takahashi, N, Ishii, S, Okura, Y, Fukami, T.A, Torizawa, T. | | Deposit date: | 2023-08-31 | | Release date: | 2023-11-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.236 Å) | | Cite: | Characterizations of a neutralizing antibody broadly reactive to multiple gluten peptide:HLA-DQ2.5 complexes in the context of celiac disease.
Nat Commun, 14, 2023
|
|
8W84
 
 | | HLA-DQ2.5-alpha2 gliadin peptide in complex with DQN0344AE02 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DQN0344AE02 Fab heavy chain, DQN0344AE02 Fab light chain, ... | | Authors: | Irie, M, Tsushima, T, Teranishi-Ikawa, Y, Takahashi, N, Ishii, S, Okura, Y, Fukami, T.A, Torizawa, T. | | Deposit date: | 2023-08-31 | | Release date: | 2023-11-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.105 Å) | | Cite: | Characterizations of a neutralizing antibody broadly reactive to multiple gluten peptide:HLA-DQ2.5 complexes in the context of celiac disease.
Nat Commun, 14, 2023
|
|
2PCK
 
 | | Crystal structure of PH0725 from Pyrococcus horikoshii OT3 | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, diphthine synthase | | Authors: | Yamamoto, H, Morikawa, Y, Matsuura, Y, Shimada, H, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-30 | | Release date: | 2007-10-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of PH0725 from Pyrococcus horikoshii OT3
To be Published
|
|
2E4R
 
 | |
2EH4
 
 | |
2ED5
 
 | |
2P6I
 
 | | Crystal structure of PH0725 from Pyrococcus horikoshii OT3 | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, diphthine synthase | | Authors: | Yamamoto, H, Matsuura, Y, Morikawa, Y, Shimada, H, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-18 | | Release date: | 2007-09-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of PH0725 from Pyrococcus horikoshii OT3
To be Published
|
|
3AM2
 
 | | Clostridium perfringens enterotoxin | | Descriptor: | GLYCEROL, Heat-labile enterotoxin B chain, UNKNOWN ATOM OR ION | | Authors: | Kitadokoro, K, Nishimura, K, Kamitani, S, Kimura, J, Fukui, A, Abe, H, Horiguchi, Y. | | Deposit date: | 2010-08-12 | | Release date: | 2011-04-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystal Structure of Clostridium perfringens Enterotoxin Displays Features of {beta}-Pore-forming Toxins
J.Biol.Chem., 286, 2011
|
|
3AC9
 
 | |
8RKI
 
 | | Molecular basis of ZP3/ZP1 heteropolymerization: crystal structure of a native vertebrate egg coat filament fragment | | Descriptor: | Choriogenin H, YTTERBIUM (III) ION, Zona pellucida sperm-binding protein 3, ... | | Authors: | Wiseman, B, Zamora-Caballero, S, de Sanctis, D, Yasumasu, S, Jovine, L. | | Deposit date: | 2023-12-25 | | Release date: | 2024-03-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | ZP2 cleavage blocks polyspermy by modulating the architecture of the egg coat.
Cell, 187, 2024
|
|
