7TRA
 
 | | Cascade complex from type I-A CRISPR-Cas system | | Descriptor: | CRISPR-associated endonuclease Cas3-HD, CRISPR-associated helicase Cas3, Cas11a, ... | | Authors: | Hu, C, Ni, D, Nam, K.H, Majumdar, S, McLean, J, Stahlberg, H, Terns, M, Ke, A. | | Deposit date: | 2022-01-28 | | Release date: | 2022-08-10 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Allosteric control of type I-A CRISPR-Cas3 complexes and establishment as effective nucleic acid detection and human genome editing tools.
Mol.Cell, 82, 2022
|
|
7TR9
 
 | | Cascade complex from type I-A CRISPR-Cas system | | Descriptor: | CRISPR-associated endonuclease Cas3-HD, CRISPR-associated helicase Cas3, Cas11a, ... | | Authors: | Hu, C, Ni, D, Nam, K.H, Majumdar, S, McLean, J, Stahlberg, H, Terns, M, Ke, A. | | Deposit date: | 2022-01-28 | | Release date: | 2022-08-10 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Allosteric control of type I-A CRISPR-Cas3 complexes and establishment as effective nucleic acid detection and human genome editing tools.
Mol.Cell, 82, 2022
|
|
5Y8Q
 
 | | ZsYellow at pH 8.0 | | Descriptor: | GFP-like fluorescent chromoprotein FP538 | | Authors: | Bae, J.E, Kim, I.J, Nam, K.H. | | Deposit date: | 2017-08-21 | | Release date: | 2017-09-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Disruption of the hydrogen bonding network determines the pH-induced non-fluorescent state of the fluorescent protein ZsYellow by protonation of Glu221.
Biochem. Biophys. Res. Commun., 493, 2017
|
|
5Y4J
 
 | |
5Y4I
 
 | | Crystal structure of glucose isomerase in complex with glycerol in one metal binding mode | | Descriptor: | ACETATE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Bae, J.E, Kim, I.J, Nam, K.H. | | Deposit date: | 2017-08-03 | | Release date: | 2017-09-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal structure of glucose isomerase in complex with xylitol inhibitor in one metal binding mode
Biochem. Biophys. Res. Commun., 493, 2017
|
|
5Z6V
 
 | |
6AA7
 
 | |
5XVN
 
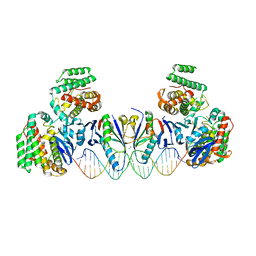 | | E. far Cas1-Cas2/prespacer binary complex | | Descriptor: | CRISPR-associated endonuclease Cas1, CRISPR-associated endoribonuclease Cas2, DNA (28-MER), ... | | Authors: | Xiao, Y, Ng, S, Nam, K.H, Ke, A. | | Deposit date: | 2017-06-28 | | Release date: | 2017-10-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | How type II CRISPR-Cas establish immunity through Cas1-Cas2-mediated spacer integration.
Nature, 550, 2017
|
|
5XVO
 
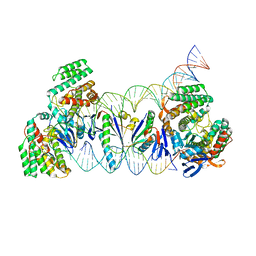 | | E. fae Cas1-Cas2/prespacer/target ternary complex revealing DNA sampling and half-integration states | | Descriptor: | CRISPR-associated endonuclease Cas1, CRISPR-associated endoribonuclease Cas2, DNA (28-MER), ... | | Authors: | Xiao, Y, Ng, S, Nam, K.H, Ke, A. | | Deposit date: | 2017-06-28 | | Release date: | 2017-10-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | How type II CRISPR-Cas establish immunity through Cas1-Cas2-mediated spacer integration.
Nature, 550, 2017
|
|
5XVP
 
 | | E. fae Cas1-Cas2/prespacer/target ternary complex revealing the fully integrated states | | Descriptor: | CRISPR-associated endonuclease Cas1, CRISPR-associated endoribonuclease Cas2, DNA (5'-D(P*TP*TP*CP*TP*CP*CP*GP*AP*G)-3'), ... | | Authors: | Xiao, Y, Ng, S, Nam, K.H, Ke, A. | | Deposit date: | 2017-06-28 | | Release date: | 2017-10-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | How type II CRISPR-Cas establish immunity through Cas1-Cas2-mediated spacer integration.
Nature, 550, 2017
|
|
5Y9Q
 
 | |
7MID
 
 | | Sub-complex of Cas4-Cas1-Cas2 bound PAM containing DNA | | Descriptor: | CRISPR-associated endoribonuclease Cas2, CRISPR-associated exonuclease Cas4/endonuclease Cas1 fusion, DNA (33-MER), ... | | Authors: | Hu, C.Y, Ke, A.K. | | Deposit date: | 2021-04-16 | | Release date: | 2021-11-17 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Mechanism for Cas4-assisted directional spacer acquisition in CRISPR-Cas.
Nature, 598, 2021
|
|
7MI4
 
 | | Symmetrical PAM-PAM prespacer bound Cas4/Cas1/Cas2 complex | | Descriptor: | CRISPR-associated endoribonuclease Cas2, CRISPR-associated exonuclease Cas4/endonuclease Cas1 fusion, DNA (35-MER), ... | | Authors: | Hu, C.Y, Ke, A.K. | | Deposit date: | 2021-04-16 | | Release date: | 2021-11-17 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanism for Cas4-assisted directional spacer acquisition in CRISPR-Cas.
Nature, 598, 2021
|
|
7MI5
 
 | | Asymmetrical PAM-Non PAM prespacer bound Cas4/Cas1/Cas2 complex | | Descriptor: | CRISPR-associated endoribonuclease Cas2, CRISPR-associated exonuclease Cas4/endonuclease Cas1 fusion, DNA (26-MER), ... | | Authors: | Hu, C.Y, Ke, A.K. | | Deposit date: | 2021-04-16 | | Release date: | 2021-11-17 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Mechanism for Cas4-assisted directional spacer acquisition in CRISPR-Cas.
Nature, 598, 2021
|
|
7MI9
 
 | | Full integration complex of Cas1/Cas2 from Cas4-containing system | | Descriptor: | CRISPR-associated endoribonuclease Cas2, CRISPR-associated exonuclease Cas4/endonuclease Cas1 fusion, DNA (5'-D(P*CP*GP*GP*AP*AP*AP*AP*GP*AP*GP*CP*C)-3'), ... | | Authors: | Hu, C.Y, Ke, A.K. | | Deposit date: | 2021-04-16 | | Release date: | 2021-11-17 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.89 Å) | | Cite: | Mechanism for Cas4-assisted directional spacer acquisition in CRISPR-Cas.
Nature, 598, 2021
|
|
7MIB
 
 | |
2OHE
 
 | | Structural and mutational analysis of tRNA-Intron splicing endonuclease from Thermoplasma acidophilum DSM 1728 | | Descriptor: | tRNA-splicing endonuclease | | Authors: | Kim, Y.K, Mizutani, K, Rhee, K.H, Lee, W.H, Park, S.Y, Hwang, K.Y. | | Deposit date: | 2007-01-10 | | Release date: | 2007-11-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Mutational Analysis of tRNA Intron-Splicing Endonuclease from Thermoplasma acidophilum DSM 1728: Catalytic Mechanism of tRNA Intron-Splicing Endonucleases
J.Bacteriol., 189, 2007
|
|
2OHC
 
 | | structural and mutational analysis of tRNA-intron splicing endonuclease from Thermoplasma acidophilum DSM1728 | | Descriptor: | tRNA-splicing endonuclease | | Authors: | Kim, Y.K, Mizutani, K, Rhee, K.H, Lee, W.H, Park, S.Y, Hwang, K.Y. | | Deposit date: | 2007-01-10 | | Release date: | 2007-11-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Mutational Analysis of tRNA Intron-Splicing Endonuclease from Thermoplasma acidophilum DSM 1728: Catalytic Mechanism of tRNA Intron-Splicing Endonucleases
J.Bacteriol., 189, 2007
|
|
7YGF
 
 | | Crystal structure of YggS from Fusobacterium nucleatum | | Descriptor: | Pyridoxal phosphate homeostasis protein, SULFATE ION | | Authors: | He, S.R, Chan, Y.Y, Wang, L.L, Bai, X, Bu, T.T, Zhang, J, Xu, Y.B. | | Deposit date: | 2022-07-11 | | Release date: | 2022-10-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural and Functional Analysis of the Pyridoxal Phosphate Homeostasis Protein YggS from Fusobacterium nucleatum.
Molecules, 27, 2022
|
|
2NX8
 
 | |
3E6E
 
 | | Crystal structure of Alanine racemase from E.faecalis complex with cycloserine | | Descriptor: | Alanine racemase, D-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL]-N,O-CYCLOSERYLAMIDE | | Authors: | Hwang, K.Y, Priyadarshi, A, Lee, E.H, Sung, M.W. | | Deposit date: | 2008-08-15 | | Release date: | 2009-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into the alanine racemase from Enterococcus faecalis.
Biochim.Biophys.Acta, 1794, 2009
|
|
3DRE
 
 | |
3DRB
 
 | | Crystal structure of Human Brain-type Creatine Kinase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Creatine kinase B-type, MAGNESIUM ION | | Authors: | Moon, J.H, Bong, S.M, Hwang, K.Y, Chi, Y.M. | | Deposit date: | 2008-07-11 | | Release date: | 2009-03-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural studies of human brain-type creatine kinase complexed with the ADP-Mg2+-NO3- -creatine transition-state analogue complex
Febs Lett., 582, 2008
|
|
3FLM
 
 | | Crystal structure of menD from E.coli | | Descriptor: | 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-1-carboxylate synthase | | Authors: | Priyadarshi, A, Hwang, K.Y. | | Deposit date: | 2008-12-19 | | Release date: | 2009-03-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insights of the MenD from Escherichia coli reveal ThDP affinity.
Biochem.Biophys.Res.Commun., 380, 2009
|
|
3FTJ
 
 | |
