7DAW
 
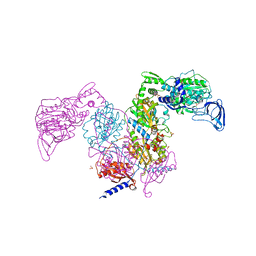 | | Crystal structure of Mycobacterium tuberculosis phenylalanyl-tRNA synthetase | | Descriptor: | Phenylalanine--tRNA ligase alpha subunit, Phenylalanine--tRNA ligase beta subunit, SULFATE ION | | Authors: | Xu, M, Zhang, X, Xu, L, Chen, S. | | Deposit date: | 2020-10-18 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Re-discovery of PF-3845 as a new chemical scaffold inhibiting phenylalanyl-tRNA synthetase in Mycobacterium tuberculosis .
J.Biol.Chem., 2021
|
|
7DB8
 
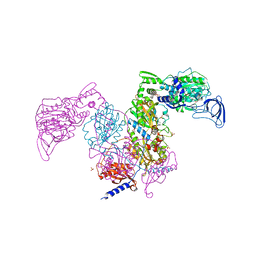 | | Crystal structure of Mycobacterium tuberculosis phenylalanyl-tRNA synthetase in complex with compound PF-3845 | | Descriptor: | N-pyridin-3-yl-4-[[3-[5-(trifluoromethyl)pyridin-2-yl]oxyphenyl]methyl]piperidine-1-carboxamide, Phenylalanine--tRNA ligase alpha subunit, Phenylalanine--tRNA ligase beta subunit, ... | | Authors: | Xu, M, Zhang, X, Xu, L, Chen, S. | | Deposit date: | 2020-10-19 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Re-discovery of PF-3845 as a new chemical scaffold inhibiting phenylalanyl-tRNA synthetase in Mycobacterium tuberculosis .
J.Biol.Chem., 2021
|
|
7E40
 
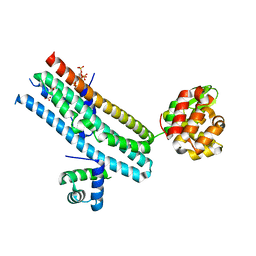 | | Mechanism of Phosphate Sensing and Signaling Revealed by Rice SPX1-PHR2 Complex Structure | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, Protein PHOSPHATE STARVATION RESPONSE 2, SPX domain-containing protein 1,Endolysin | | Authors: | Zhou, J, Hu, Q, Yao, D, Xing, W. | | Deposit date: | 2021-02-09 | | Release date: | 2021-11-10 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mechanism of phosphate sensing and signaling revealed by rice SPX1-PHR2 complex structure.
Nat Commun, 12, 2021
|
|
8INB
 
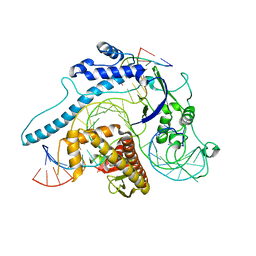 | | Cryo-EM structure of Cas12j-SF05-crRNA-dsDNA complex | | Descriptor: | Cas12j-SF05, NTS-DNA, TS-DNA, ... | | Authors: | Zhang, X, Duan, Z.Q, Zhu, J.K. | | Deposit date: | 2023-03-09 | | Release date: | 2024-03-13 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular basis for DNA cleavage by the hypercompact Cas12j-SF05.
Cell Discov, 9, 2023
|
|
7YNK
 
 | |
7YNJ
 
 | |
7YNI
 
 | | Structure of human SGLT1-MAP17 complex bound with substrate 4D4FDG in the occluded conformation | | Descriptor: | (2R,3R,4R,5S,6R)-5-fluoranyl-6-(hydroxymethyl)oxane-2,3,4-triol, PDZK1-interacting protein 1, Sodium/glucose cotransporter 1 | | Authors: | Chen, L, Niu, Y, Cui, W. | | Deposit date: | 2022-07-31 | | Release date: | 2023-05-31 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structures of human SGLT in the occluded state reveal conformational changes during sugar transport.
Nat Commun, 14, 2023
|
|
8K8K
 
 | | Structure of Klebsiella pneumonia ModA | | Descriptor: | Molybdate transporter periplasmic protein | | Authors: | Zhao, Q, Bartlam, M. | | Deposit date: | 2023-07-31 | | Release date: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural analysis of molybdate binding protein ModA from Klebsiella pneumoniae.
Biochem.Biophys.Res.Commun., 681, 2023
|
|
8K8L
 
 | |
7YT9
 
 | | crystal structure of AGD1-4 of Arabidopsis AGDP3 | | Descriptor: | AGD1-4 of Arabidopsis AGDP3 | | Authors: | Zhou, X, Du, J. | | Deposit date: | 2022-08-13 | | Release date: | 2022-10-12 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The H3K9me2-binding protein AGDP3 limits DNA methylation and transcriptional gene silencing in Arabidopsis.
J Integr Plant Biol, 64, 2022
|
|
7YTA
 
 | |
5YKR
 
 | |
5YKT
 
 | |
5Z8L
 
 | |
5ZNR
 
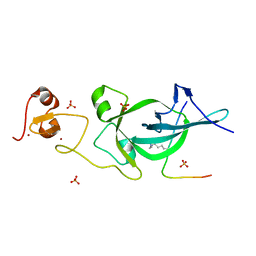 | | Crystal structure of PtSHL in complex with an H3K27me3 peptide | | Descriptor: | 17-mer peptide from Histone H3.2, SHORT LIFE family protein, SULFATE ION, ... | | Authors: | Lv, X, Du, J. | | Deposit date: | 2018-04-10 | | Release date: | 2018-07-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.202 Å) | | Cite: | Dual recognition of H3K4me3 and H3K27me3 by a plant histone reader SHL.
Nat Commun, 9, 2018
|
|
6A5M
 
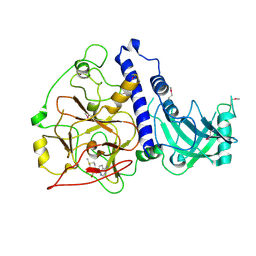 | | Crystal structure of Arabidopsis thaliana SUVH6 in complex with SAM, form 2 | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH6, S-ADENOSYLMETHIONINE, ... | | Authors: | Li, X, Du, J. | | Deposit date: | 2018-06-24 | | Release date: | 2018-08-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Mechanistic insights into plant SUVH family H3K9 methyltransferases and their binding to context-biased non-CG DNA methylation.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6A5N
 
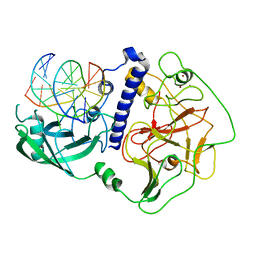 | | Crystal structure of Arabidopsis thaliana SUVH6 in complex with methylated DNA | | Descriptor: | DNA (5'-D(*CP*AP*CP*TP*GP*CP*TP*GP*AP*GP*TP*AP*CP*T)-3'), DNA (5'-D(*GP*AP*GP*TP*AP*CP*TP*(5CM)P*AP*GP*CP*AP*GP*T)-3'), Histone-lysine N-methyltransferase, ... | | Authors: | Li, X, Du, J. | | Deposit date: | 2018-06-24 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mechanistic insights into plant SUVH family H3K9 methyltransferases and their binding to context-biased non-CG DNA methylation.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6A5K
 
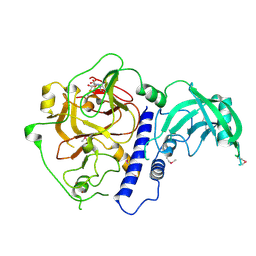 | | Crystal structure of Arabidopsis thaliana SUVH6 in complex with SAM, form 1 | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH6, S-ADENOSYLMETHIONINE, ... | | Authors: | Li, X, Du, J. | | Deposit date: | 2018-06-24 | | Release date: | 2018-08-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanistic insights into plant SUVH family H3K9 methyltransferases and their binding to context-biased non-CG DNA methylation.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5Z8N
 
 | |
5ZNP
 
 | | Crystal structure of PtSHL in complex with an H3K4me3 peptide | | Descriptor: | 15-mer peptide from Histone H3.2, SHORT LIFE family protein, ZINC ION | | Authors: | Lv, X, Du, J. | | Deposit date: | 2018-04-10 | | Release date: | 2018-07-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Dual recognition of H3K4me3 and H3K27me3 by a plant histone reader SHL.
Nat Commun, 9, 2018
|
|
8Y1J
 
 | |
8XTF
 
 | | Crystal structure of methyltransferase MpaG' in complex with SAH and FDHMP-3C | | Descriptor: | 4-farnesyl-3,5-dihydroxy-6-methylphthalide-3C, O-methyltransferase mpaG', S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | You, C, Pan, Y.J, Li, S.Y, Feng, Y.G. | | Deposit date: | 2024-01-10 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structural basis for substrate flexibility of the O-methyltransferase MpaG' involved in mycophenolic acid biosynthesis.
Protein Sci., 33, 2024
|
|
8XTG
 
 | | Crystal structure of methyltransferase MpaG' in complex with SAH and DMMPA | | Descriptor: | O-desmethyl mycophenolic acid, O-methyltransferase mpaG', S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | You, C, Pan, Y.J, Li, S.Y, Feng, Y.G. | | Deposit date: | 2024-01-10 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for substrate flexibility of the O-methyltransferase MpaG' involved in mycophenolic acid biosynthesis.
Protein Sci., 33, 2024
|
|
8XTE
 
 | | Crystal structure of methyltransferase MpaG' in complex with SAH and FDHMP | | Descriptor: | 4-farnesyl-3,5-dihydroxy-6-methylphthalide, O-methyltransferase mpaG', S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | You, C, Pan, Y.J, Li, S.Y, Feng, Y.G. | | Deposit date: | 2024-01-10 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural basis for substrate flexibility of the O-methyltransferase MpaG' involved in mycophenolic acid biosynthesis.
Protein Sci., 33, 2024
|
|
6IN8
 
 | | Crystal structure of MucB | | Descriptor: | Sigma factor AlgU regulatory protein MucB | | Authors: | Li, S, Zhang, Q, Bartlam, M. | | Deposit date: | 2018-10-24 | | Release date: | 2019-07-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the recognition of MucA by MucB and AlgU in Pseudomonas aeruginosa.
Febs J., 286, 2019
|
|
