8R1D
 
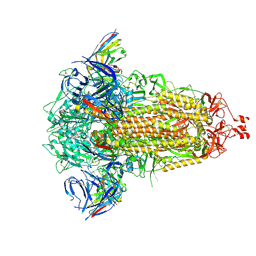 | | SD1-3 Fab in complex with SARS-CoV-2 BA.2.12.1 Spike Glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SD1-3 Fab Heavy Chain, SD1-3 Fab Light Chain, ... | | Authors: | Duyvesteyn, H.M.E, Ren, J, Stuart, D.I. | | Deposit date: | 2023-11-01 | | Release date: | 2024-03-13 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.37 Å) | | Cite: | The SARS-CoV-2 neutralizing antibody response to SD1 and its evasion by BA.2.86.
Nat Commun, 15, 2024
|
|
8QZR
 
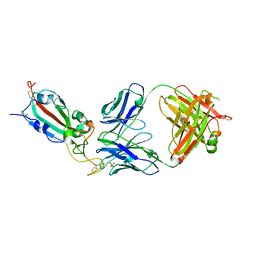 | | SARS-CoV-2 delta RBD complexed with BA.4/5-9 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BA.4/5-9 heavy chain, BA.4/5-9 light chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2023-10-29 | | Release date: | 2024-04-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.77 Å) | | Cite: | Emerging variants develop total escape from potent monoclonal antibodies induced by BA.4/5 infection.
Nat Commun, 15, 2024
|
|
8R80
 
 | | SARS-CoV-2 Delta RBD in complex with XBB-9 Fab and an anti-Fab nanobody | | Descriptor: | Spike protein S1, XBB-9 Fab heavy chain, XBB-9 Fab light chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2023-11-27 | | Release date: | 2024-05-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (4.03 Å) | | Cite: | A structure-function analysis shows SARS-CoV-2 BA.2.86 balances antibody escape and ACE2 affinity.
Cell Rep Med, 5, 2024
|
|
8QRF
 
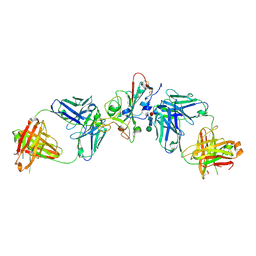 | | SARS-CoV-2 delta RBD complexed with XBB-6 and beta-49 Fabs | | Descriptor: | Beta-49 heavy chain, Beta-49 light chain, Spike protein S1, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2023-10-06 | | Release date: | 2024-05-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | A structure-function analysis shows SARS-CoV-2 BA.2.86 balances antibody escape and ACE2 affinity.
Cell Rep Med, 5, 2024
|
|
8QTD
 
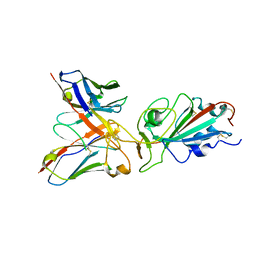 | | Local refinement of SARS-CoV-2 BA.2.86 Spike and XBB-7 Fab | | Descriptor: | Spike glycoprotein,Fibritin, XBB-7 fab heavy chain, XBB-7 fab light chain | | Authors: | Ren, J, Duyvesteyn, H.M.E, Stuart, D.I. | | Deposit date: | 2023-10-12 | | Release date: | 2024-05-08 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | A structure-function analysis shows SARS-CoV-2 BA.2.86 balances antibody escape and ACE2 affinity.
Cell Rep Med, 5, 2024
|
|
8QRG
 
 | | SARS-CoV-2 delta RBD complexed with XBB-2 Fab and NbC1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, NbC1, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2023-10-07 | | Release date: | 2024-05-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A structure-function analysis shows SARS-CoV-2 BA.2.86 balances antibody escape and ACE2 affinity.
Cell Rep Med, 5, 2024
|
|
8R8K
 
 | | XBB-4 Fab in complex with SARS-CoV-2 BA.2.12.1 Spike Glycoprotein | | Descriptor: | Spike glycoprotein,Fibritin, XBB-4 Fab Heavy chain, XBB-4 Fab Light chain | | Authors: | Duyvesteyn, H.M.E, Ren, J, Stuart, D.I. | | Deposit date: | 2023-11-29 | | Release date: | 2024-05-08 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | A structure-function analysis shows SARS-CoV-2 BA.2.86 balances antibody escape and ACE2 affinity.
Cell Rep Med, 5, 2024
|
|
5OVQ
 
 | |
9L0Q
 
 | |
9HGD
 
 | | Crystal structure of human GABARAP in complex with cyclic peptide GAB_D23 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Ueffing, A, Wilms, J.A, Willbold, D, Weiergraeber, O.H. | | Deposit date: | 2024-11-19 | | Release date: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Accurate de novo design of high-affinity protein-binding macrocycles using deep learning.
Nat.Chem.Biol., 2025
|
|
8HNK
 
 | | CXCR3-DNGi complex activated by CXCL11 | | Descriptor: | C-X-C motif chemokine 11, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Jiao, H.Z, Hu, H.L. | | Deposit date: | 2022-12-08 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural insights into the activation and inhibition of CXC chemokine receptor 3.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8HNN
 
 | | Structure of CXCR3 complexed with antagonist SCH546738 | | Descriptor: | 3-azanyl-6-chloranyl-5-[(3S)-4-[1-[(4-chlorophenyl)methyl]piperidin-4-yl]-3-ethyl-piperazin-1-yl]pyrazine-2-carboxamide, CHOLESTEROL, Nb6, ... | | Authors: | Jiao, H.Z, Hu, H.L. | | Deposit date: | 2022-12-08 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural insights into the activation and inhibition of CXC chemokine receptor 3.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8HNL
 
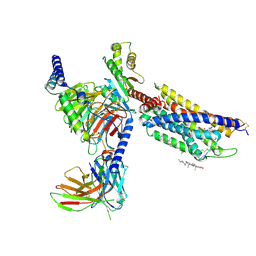 | | CXCR3-DNGi complex activated by PS372424 | | Descriptor: | (3S)-N-[(2S)-5-carbamimidamido-1-(cyclohexylmethylamino)-1-oxidanylidene-pentan-2-yl]-2-(4-oxidanylidene-4-phenyl-butanoyl)-3,4-dihydro-1H-isoquinoline-3-carboxamide, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Jiao, H.Z, Hu, H.L. | | Deposit date: | 2022-12-08 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structural insights into the activation and inhibition of CXC chemokine receptor 3.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8HNM
 
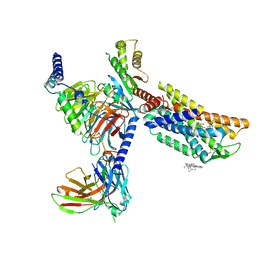 | | CXCR3-DNGi complex activated by VUF11222 | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Jiao, H.Z, Hu, H.L. | | Deposit date: | 2022-12-08 | | Release date: | 2023-11-29 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Structural insights into the activation and inhibition of CXC chemokine receptor 3.
Nat.Struct.Mol.Biol., 31, 2024
|
|
9HGC
 
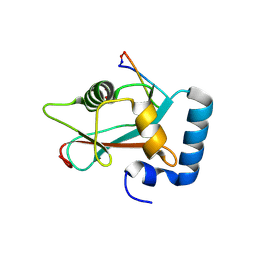 | |
7FEC
 
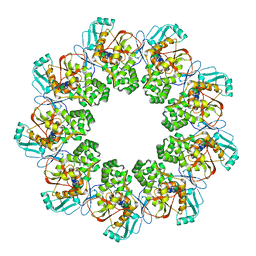 | |
7FED
 
 | |
7FEB
 
 | |
9JDC
 
 | | Structure of chanoclavine synthase from Claviceps fusiformis in complex with prechanoclavine | | Descriptor: | (2~{S})-2-(methylamino)-3-[4-[(1~{E})-3-methylbuta-1,3-dienyl]-1~{H}-indol-3-yl]propanoic acid, Catalase easC, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Liu, Z.W, Wang, T, Li, X, Shen, P.P, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2024-08-31 | | Release date: | 2025-01-01 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (2.33 Å) | | Cite: | Chanoclavine synthase operates by an NADPH-independent superoxide mechanism.
Nature, 640, 2025
|
|
9JDB
 
 | | Structure of chanoclavine synthase from Claviceps fusiformis | | Descriptor: | Catalase easC, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Liu, Z.W, Wang, T, Li, X, Shen, P.P, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2024-08-31 | | Release date: | 2025-01-01 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Chanoclavine synthase operates by an NADPH-independent superoxide mechanism.
Nature, 640, 2025
|
|
1Z1Z
 
 | |
8KEI
 
 | | Cryo-EM structure of NADPH oxidase 2 in complex with p22phox and EROS | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Liang, S.Y, Liu, A.J, Liu, Y.Z, Ye, R.D. | | Deposit date: | 2023-08-11 | | Release date: | 2024-05-22 | | Last modified: | 2024-12-04 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Structural basis for EROS binding to human phagocyte NADPH oxidase NOX2.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8K3Z
 
 | | Cryo-EM structure of CXCR4 in complex with CXCL12 | | Descriptor: | C-X-C chemokine receptor type 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, Y.Z, Liu, A.J, Liao, Q.W, Ye, R.D. | | Deposit date: | 2023-07-17 | | Release date: | 2024-07-17 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Cryo-EM structure of monomeric CXCL12-bound CXCR4 in the active state.
Cell Rep, 43, 2024
|
|
4FA1
 
 | |
4FAN
 
 | |
