8GHU
 
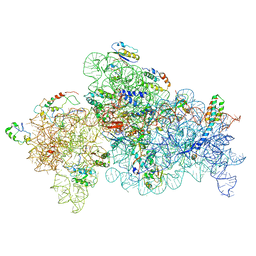 | |
8F5K
 
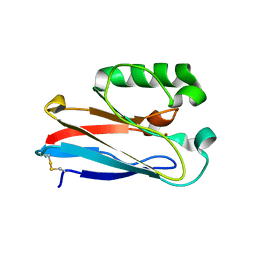 | |
8F5L
 
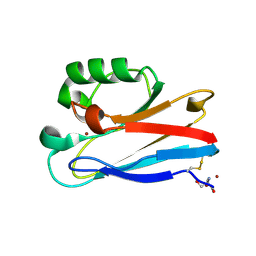 | | Azurin from Pseudomonas aeruginosa, Y72F/Y108F/F110L mutant | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Azurin, COPPER (II) ION | | Authors: | Zeug, M, Offenbacher, A.R, Choe, J. | | Deposit date: | 2022-11-14 | | Release date: | 2023-01-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Electrochemical and Structural Study of the Buried Tryptophan in Azurin: Effects of Hydration and Polarity on the Redox Potential of W48.
J.Phys.Chem.B, 127, 2023
|
|
6FR2
 
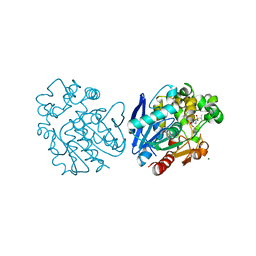 | | Soluble epoxide hydrolase in complex with LK864 | | Descriptor: | 1-[(4~{S})-9-propan-2-ylsulfonyl-1-oxa-9-azaspiro[5.5]undecan-4-yl]-3-[[4-(trifluoromethyloxy)phenyl]methyl]urea, Bifunctional epoxide hydrolase 2, MAGNESIUM ION | | Authors: | Kramer, J.S, Pogoryelov, D, Krasavin, M, Proschak, E. | | Deposit date: | 2018-02-15 | | Release date: | 2018-09-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.262 Å) | | Cite: | Discovery of polar spirocyclic orally bioavailable urea inhibitors of soluble epoxide hydrolase.
Bioorg. Chem., 80, 2018
|
|
6YLC
 
 | | Biochemical, Cellular and Structural Characterization of Novel ERK3 Inhibitors | | Descriptor: | 5-fluoranyl-2-[5-[[1-(1-methylpiperidin-4-yl)pyrazol-4-yl]amino]-[1,2,3]triazolo[4,5-d]pyrimidin-3-yl]benzenecarbonitrile, Mitogen-activated protein kinase 6 | | Authors: | Graedler, U. | | Deposit date: | 2020-04-07 | | Release date: | 2020-09-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Biochemical, cellular and structural characterization of novel and selective ERK3 inhibitors.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
6YKY
 
 | | Biochemical, Cellular and Structural Characterization of Novel ERK3 Inhibitors | | Descriptor: | 3-(4-methoxyphenyl)-~{N}-[(3~{R})-1-pyridin-4-ylpyrrolidin-3-yl]-[1,2,3]triazolo[4,5-d]pyrimidin-5-amine, Mitogen-activated protein kinase 6 | | Authors: | Graedler, U. | | Deposit date: | 2020-04-06 | | Release date: | 2020-09-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Biochemical, cellular and structural characterization of novel and selective ERK3 inhibitors.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
6YLL
 
 | | Biochemical, Cellular and Structural Characterization of Novel ERK3 Inhibitors | | Descriptor: | Mitogen-activated protein kinase 6, ~{N}4-[3-(4-methoxyphenyl)-[1,2,3]triazolo[4,5-d]pyrimidin-5-yl]cyclohexane-1,4-diamine | | Authors: | Graedler, U. | | Deposit date: | 2020-04-07 | | Release date: | 2020-09-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Biochemical, cellular and structural characterization of novel and selective ERK3 inhibitors.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
5I5S
 
 | |
5I5V
 
 | | X-RAY CRYSTAL STRUCTURE AT 1.94A RESOLUTION OF HUMAN MITOCHONDRIAL BRANCHED CHAIN AMINOTRANSFERASE (BCATM) COMPLEXED WITH A THIENOPYRIMIDINE COMPOUND AND AN INTERNAL ALDIMINE LINKED PLP COFACTOR. | | Descriptor: | 1,2-ETHANEDIOL, 3,5-dimethyl-4-oxo-3,4-dihydrothieno[2,3-d]pyrimidine-6-carboxylic acid, Branched-chain-amino-acid aminotransferase, ... | | Authors: | Somers, D.O. | | Deposit date: | 2016-02-15 | | Release date: | 2016-03-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structurally Diverse Mitochondrial Branched Chain Aminotransferase (BCATm) Leads with Varying Binding Modes Identified by Fragment Screening.
J.Med.Chem., 59, 2016
|
|
5I5Y
 
 | |
5I60
 
 | |
6SLG
 
 | | HUMAN ERK2 WITH ERK1/2 INHIBITOR, AZD0364. | | Descriptor: | (6~{R})-7-[[3,4-bis(fluoranyl)phenyl]methyl]-6-(methoxymethyl)-2-[5-methyl-2-[(2-methylpyrazol-3-yl)amino]pyrimidin-4-yl]-5,6-dihydroimidazo[1,2-a]pyrazin-8-one, 1,2-ETHANEDIOL, ERK-tide, ... | | Authors: | Breed, J, Phillips, C. | | Deposit date: | 2019-08-19 | | Release date: | 2019-11-20 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Discovery of a Potent and Selective Oral Inhibitor of ERK1/2 (AZD0364) That Is Efficacious in Both Monotherapy and Combination Therapy in Models of Nonsmall Cell Lung Cancer (NSCLC).
J.Med.Chem., 62, 2019
|
|
5I5U
 
 | | X-RAY CRYSTAL STRUCTURE AT 2.40A RESOLUTION OF HUMAN MITOCHONDRIAL BRANCHED CHAIN AMINOTRANSFERASE (BCATM) COMPLEXED WITH A TETRAHYDRONAPHTHALENYL COMPOUND AND AN INTERNAL ALDIMINE LINKED PLP COFACTOR. | | Descriptor: | 1,2-ETHANEDIOL, 2-hydroxy-N-[(1R)-1,2,3,4-tetrahydronaphthalen-1-yl]acetamide, Branched-chain-amino-acid aminotransferase, ... | | Authors: | Somers, D.O. | | Deposit date: | 2016-02-15 | | Release date: | 2016-03-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.404 Å) | | Cite: | Structurally Diverse Mitochondrial Branched Chain Aminotransferase (BCATm) Leads with Varying Binding Modes Identified by Fragment Screening.
J.Med.Chem., 59, 2016
|
|
5I5X
 
 | | X-RAY CRYSTAL STRUCTURE AT 1.65A RESOLUTION OF HUMAN MITOCHONDRIAL BRANCHED CHAIN AMINOTRANSFERASE (BCATM) COMPLEXED WITH A THIAZOLE COMPOUND AND PMP COFACTOR. | | Descriptor: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 5-methyl-4-oxo-N-(1,3,4-thiadiazol-2-yl)-3,4-dihydrothieno[2,3-d]pyrimidine-6-carboxamide, ... | | Authors: | Somers, D.O. | | Deposit date: | 2016-02-15 | | Release date: | 2016-03-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structurally Diverse Mitochondrial Branched Chain Aminotransferase (BCATm) Leads with Varying Binding Modes Identified by Fragment Screening.
J.Med.Chem., 59, 2016
|
|
5I5T
 
 | | X-RAY CRYSTAL STRUCTURE AT 2.31A RESOLUTION OF HUMAN MITOCHONDRIAL BRANCHED CHAIN AMINOTRANSFERASE (BCATM) COMPLEXED WITH A TETRAHYDROQUINOLINE COMPOUND AND AN INTERNAL ALDIMINE LINKED PLP COFACTOR. | | Descriptor: | (3R)-3-methyl-1,2,3,4-tetrahydroquinoline-8-sulfonamide, 1,2-ETHANEDIOL, Branched-chain-amino-acid aminotransferase, ... | | Authors: | Somers, D.O. | | Deposit date: | 2016-02-15 | | Release date: | 2016-03-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structurally Diverse Mitochondrial Branched Chain Aminotransferase (BCATm) Leads with Varying Binding Modes Identified by Fragment Screening.
J.Med.Chem., 59, 2016
|
|
5I5W
 
 | |
5CR5
 
 | |
7PTQ
 
 | | RNA origami 5-helix tile | | Descriptor: | Chains: C | | Authors: | McRae, E.K.S, Andersen, E.S. | | Deposit date: | 2021-09-27 | | Release date: | 2022-10-05 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.08 Å) | | Cite: | Structure, folding and flexibility of co-transcriptional RNA origami.
Nat Nanotechnol, 18, 2023
|
|
7PTS
 
 | | RNA origami 5-helix tile | | Descriptor: | 5HT-B | | Authors: | McRae, E.K.S, Andersen, E.S. | | Deposit date: | 2021-09-27 | | Release date: | 2022-10-05 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (5.71 Å) | | Cite: | Structure, folding and flexibility of co-transcriptional RNA origami.
Nat Nanotechnol, 18, 2023
|
|
7PTL
 
 | |
7PTK
 
 | |
7PI6
 
 | | Trypanosoma brucei ISG65 bound to human complement C3d | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 65 kDa invariant surface glycoprotein, Complement C3dg fragment, ... | | Authors: | Cook, A.D, Higgins, M.K. | | Deposit date: | 2021-08-19 | | Release date: | 2022-07-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Invariant surface glycoprotein 65 of Trypanosoma brucei is a complement C3 receptor.
Nat Commun, 13, 2022
|
|
7QDU
 
 | |
7ZW8
 
 | |
7ZY6
 
 | |
