1G5S
 
 | | CRYSTAL STRUCTURE OF HUMAN CYCLIN DEPENDENT KINASE 2 (CDK2) IN COMPLEX WITH THE INHIBITOR H717 | | Descriptor: | 2-[TRANS-(4-AMINOCYCLOHEXYL)AMINO]-6-(BENZYL-AMINO)-9-CYCLOPENTYLPURINE, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Dreyer, M.K, Borcherding, D.R, Dumont, J.A, Peet, N.P, Tsay, J.T, Wright, P.S, Bitonti, A.J, Shen, J, Kim, S.-H. | | Deposit date: | 2000-11-02 | | Release date: | 2001-11-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal structure of human cyclin-dependent kinase 2 in complex with the adenine-derived inhibitor H717.
J.Med.Chem., 44, 2001
|
|
1LST
 
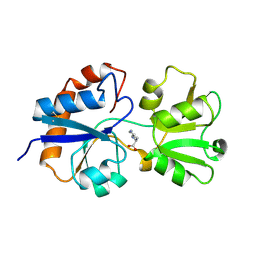 | |
1HCK
 
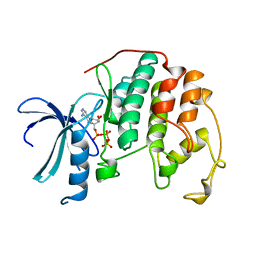 | | HUMAN CYCLIN-DEPENDENT KINASE 2 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, HUMAN CYCLIN-DEPENDENT KINASE 2, MAGNESIUM ION | | Authors: | Schulze-Gahmen, U, De Bondt, H.L, Kim, S.-H. | | Deposit date: | 1996-06-03 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-resolution crystal structures of human cyclin-dependent kinase 2 with and without ATP: bound waters and natural ligand as guides for inhibitor design.
J.Med.Chem., 39, 1996
|
|
1HCL
 
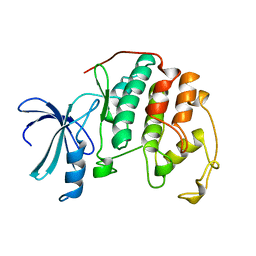 | | HUMAN CYCLIN-DEPENDENT KINASE 2 | | Descriptor: | HUMAN CYCLIN-DEPENDENT KINASE 2 | | Authors: | Schulze-Gahmen, U, De Bondt, H.L, Kim, S.-H. | | Deposit date: | 1996-06-03 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-resolution crystal structures of human cyclin-dependent kinase 2 with and without ATP: bound waters and natural ligand as guides for inhibitor design.
J.Med.Chem., 39, 1996
|
|
1NF2
 
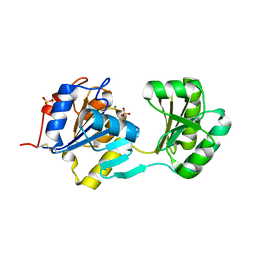 | |
1LAG
 
 | | STRUCTURAL BASES FOR MULTIPLE LIGAND SPECIFICITY OF THE PERIPLASMIC LYSINE-, ARGININE-, ORNITHINE-BINDING PROTEIN | | Descriptor: | HISTIDINE, LYSINE, ARGININE, ... | | Authors: | Kim, S.-H, Oh, B.-H. | | Deposit date: | 1993-10-06 | | Release date: | 1995-07-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural basis for multiple ligand specificity of the periplasmic lysine-, arginine-, ornithine-binding protein.
J.Biol.Chem., 269, 1994
|
|
1LAF
 
 | | STRUCTURAL BASES FOR MULTIPLE LIGAND SPECIFICITY OF THE PERIPLASMIC LYSINE-, ARGININE-, ORNITHINE-BINDING PROTEIN | | Descriptor: | ARGININE, LYSINE, ORNITHINE-BINDING PROTEIN | | Authors: | Kim, S.-H, Oh, B.-H. | | Deposit date: | 1993-10-06 | | Release date: | 1995-07-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural basis for multiple ligand specificity of the periplasmic lysine-, arginine-, ornithine-binding protein.
J.Biol.Chem., 269, 1994
|
|
2HQB
 
 | |
2HQL
 
 | |
3L0V
 
 | |
3LEA
 
 | |
3L0T
 
 | | Crystal structure of catalytic domain of TACE with hydantoin inhibitor | | Descriptor: | Disintegrin and metalloproteinase domain-containing protein 17, ISOPROPYL ALCOHOL, N-{(2R)-2-[2-(hydroxyamino)-2-oxoethyl]-4-methylpentanoyl}-3-methyl-L-valyl-N-(2-aminoethyl)-L-alaninamide, ... | | Authors: | Orth, P. | | Deposit date: | 2009-12-10 | | Release date: | 2010-03-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Discovery and SAR of hydantoin TACE inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3LE9
 
 | |
1COJ
 
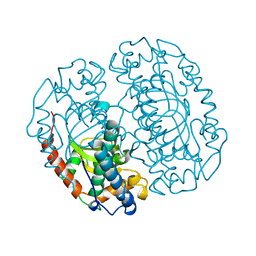 | | FE-SOD FROM AQUIFEX PYROPHILUS, A HYPERTHERMOPHILIC BACTERIUM | | Descriptor: | FE (III) ION, PROTEIN (SUPEROXIDE DISMUTASE) | | Authors: | Lim, J.H, Yu, Y.G, Kim, S.-H, Cho, S.-J, Ahn, B.Y, Han, Y.S, Cho, Y. | | Deposit date: | 1999-05-28 | | Release date: | 1999-06-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of an Fe-superoxide dismutase from the hyperthermophile Aquifex pyrophilus at 1.9 A resolution: structural basis for thermostability.
J.Mol.Biol., 270, 1997
|
|
1FJS
 
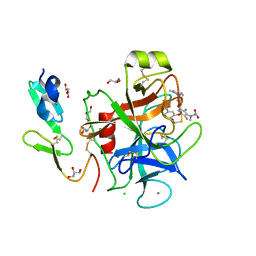 | | CRYSTAL STRUCTURE OF THE INHIBITOR ZK-807834 (CI-1031) COMPLEXED WITH FACTOR XA | | Descriptor: | CALCIUM ION, CHLORIDE ION, COAGULATION FACTOR XA, ... | | Authors: | Adler, M, Whitlow, M. | | Deposit date: | 2000-08-08 | | Release date: | 2000-10-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Preparation, characterization, and the crystal structure of the inhibitor ZK-807834 (CI-1031) complexed with factor Xa.
Biochemistry, 39, 2000
|
|
3D1N
 
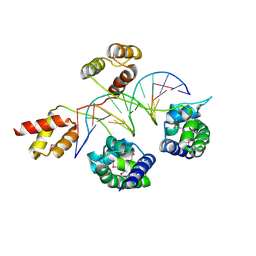 | | Structure of human Brn-5 transcription factor in complex with corticotrophin-releasing hormone gene promoter | | Descriptor: | 5'-D(*DAP*DGP*DCP*DAP*DTP*DAP*DAP*DAP*DTP*DAP*DAP*DTP*DAP*DA)-3', 5'-D(*DTP*DTP*DAP*DTP*DTP*DAP*DTP*DTP*DTP*DAP*DTP*DGP*DCP*DT)-3', POU domain, ... | | Authors: | Pereira, J.H, Ha, S.C, Kim, S.-H. | | Deposit date: | 2008-05-06 | | Release date: | 2009-05-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structure of human Brn-5 transcription factor in complex with CRH gene promoter.
J.Struct.Biol., 167, 2009
|
|
9EEZ
 
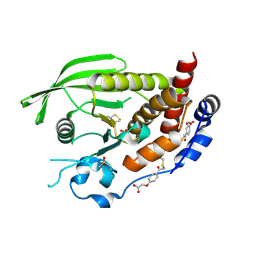 | | STEP (PTPN5) with active-site disulfide bond and covalent ligand bound to distal C505 and C518 | | Descriptor: | DIMETHYL SULFOXIDE, SULFATE ION, Tyrosine-protein phosphatase non-receptor type 5, ... | | Authors: | Guerrero, L, Ebrahim, A, Riley, B.T, Keedy, D.A. | | Deposit date: | 2024-11-19 | | Release date: | 2024-12-11 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Three STEPs forward: A trio of unexpected structures of PTPN5.
Biorxiv, 2025
|
|
9EEX
 
 | |
9EEY
 
 | |
1ZXJ
 
 | | Crystal structure of the hypthetical Mycoplasma protein, MPN555 | | Descriptor: | Hypothetical protein MG377 homolog | | Authors: | Schulze-Gahmen, U, Aono, S, Shengfeng, C, Yokota, H, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2005-06-08 | | Release date: | 2005-07-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the hypothetical Mycoplasma protein MPN555 suggests a chaperone function.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
6A6K
 
 | | Crystal structure of Estrogen-related Receptor-3 (ERR-gamma) ligand binding domain with DN201000 | | Descriptor: | 3-[(~{E})-5-oxidanyl-2-phenyl-1-[4-(4-propan-2-ylpiperazin-1-yl)phenyl]pent-1-enyl]phenol, Estrogen-related receptor gamma | | Authors: | Yoon, H, Kim, J, Chin, J, Cho, S.J, Song, J. | | Deposit date: | 2018-06-28 | | Release date: | 2019-04-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of Potent, Selective, and Orally Bioavailable Estrogen-Related Receptor-gamma Inverse Agonists To Restore the Sodium Iodide Symporter Function in Anaplastic Thyroid Cancer.
J. Med. Chem., 62, 2019
|
|
5EY2
 
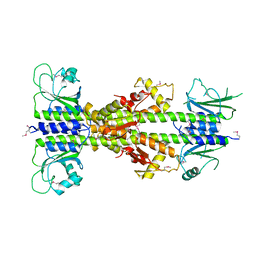 | | Crystal structure of CodY from Bacillus cereus | | Descriptor: | GTP-sensing transcriptional pleiotropic repressor CodY | | Authors: | Han, A, Lee, W.C, Son, J, Kim, S.H, Hwang, K.Y. | | Deposit date: | 2015-11-24 | | Release date: | 2016-09-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structure of the pleiotropic transcription regulator CodY provides insight into its GTP-sensing mechanism
Nucleic Acids Res., 44, 2016
|
|
1IM5
 
 | |
1STZ
 
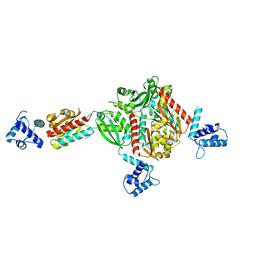 | | Crystal structure of a hypothetical protein at 2.2 A resolution | | Descriptor: | Heat-inducible transcription repressor hrcA homolog | | Authors: | Liu, J, Adams, P.D, Shin, D.-H, Huang, C, Yokota, H, Jancarik, J, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-03-25 | | Release date: | 2004-08-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a heat-inducible transcriptional repressor HrcA from Thermotoga maritima: structural insight into DNA binding and dimerization.
J.Mol.Biol., 350, 2005
|
|
1YED
 
 | |
