6IWH
 
 | | Crystal structure of rhesus macaque MHC class I molecule Mamu-B*05104 complexed with C14-GGGI lipopeptide | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, C14-GGGI lipopeptide, ... | | Authors: | Yamamoto, Y, Morita, D, Sugita, M. | | Deposit date: | 2018-12-05 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Identification and Structure of an MHC Class I-Encoded Protein with the Potential to PresentN-Myristoylated 4-mer Peptides to T Cells.
J Immunol., 202, 2019
|
|
1QRS
 
 | |
1QRT
 
 | |
1QRU
 
 | |
2RR2
 
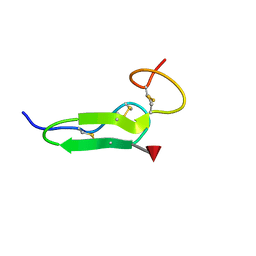 | | Structure of O-fucosylated epidermal growth factor-like repeat 12 of mouse Notch-1 receptor | | Descriptor: | Neurogenic locus notch homolog protein 1, alpha-L-fucopyranose | | Authors: | Hosoguchi, K, Shimizu, K, Fujitani, N, Nishimura, S. | | Deposit date: | 2010-02-26 | | Release date: | 2010-10-13 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Chemical Synthesis, Folding, and Structural Insights into O-Fucosylated Epidermal Growth Factor-like Repeat 12 of Mouse Notch-1 Receptor
J.Am.Chem.Soc., 132, 2010
|
|
2RR0
 
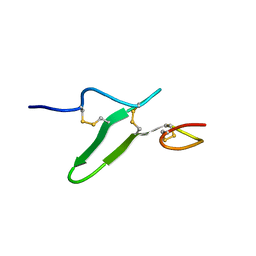 | | Structure of epidermal growth factor-like repeat 12 of mouse Notch-1 receptor | | Descriptor: | Neurogenic locus notch homolog protein 1 | | Authors: | Hosoguchi, K, Shimizu, K, Fujitani, N, Nishimura, S. | | Deposit date: | 2010-02-26 | | Release date: | 2010-10-13 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Chemical Synthesis, Folding, and Structural Insights into O-Fucosylated Epidermal Growth Factor-like Repeat 12 of Mouse Notch-1 Receptor
J.Am.Chem.Soc., 132, 2010
|
|
2RQZ
 
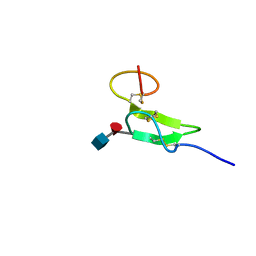 | | Structure of sugar modified epidermal growth factor-like repeat 12 of mouse Notch-1 receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-alpha-L-fucopyranose, Neurogenic locus notch homolog protein 1 | | Authors: | Shimizu, K, Fujitani, N, Hosoguchi, K, Nishimura, S. | | Deposit date: | 2010-02-26 | | Release date: | 2010-10-13 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Chemical Synthesis, Folding, and Structural Insights into O-Fucosylated Epidermal Growth Factor-like Repeat 12 of Mouse Notch-1 Receptor
J.Am.Chem.Soc., 132, 2010
|
|
1EP4
 
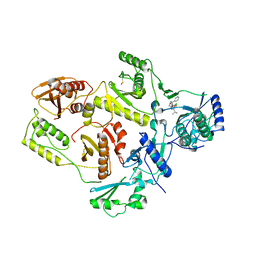 | | Crystal structure of HIV-1 reverse transcriptase in complex with S-1153 | | Descriptor: | 5-(3,5-DICHLOROPHENYL)THIO-4-ISOPROPYL-1-(PYRIDIN-4-YL-METHYL)-1H-IMIDAZOL-2-YL-METHYL CARBAMATE, HIV-1 REVERSE TRANSCRIPTASE | | Authors: | Ren, J, Nichols, C, Bird, L.E, Fujiwara, T, Suginoto, H, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2000-03-27 | | Release date: | 2000-09-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Binding of the second generation non-nucleoside inhibitor S-1153 to HIV-1 reverse transcriptase involves extensive main chain hydrogen bonding.
J.Biol.Chem., 275, 2000
|
|
5YCA
 
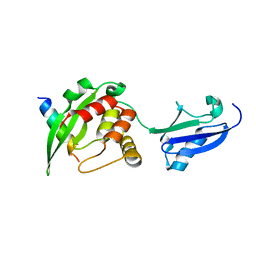 | | Crystal structure of inner membrane protein Bqt4 in complex with LEM2 | | Descriptor: | Lap-Emerin-Man domain protein 2, Ubiquitin-like protein SMT3,Bouquet formation protein 4 | | Authors: | Chen, Y, Hu, C. | | Deposit date: | 2017-09-07 | | Release date: | 2018-11-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structural insights into chromosome attachment to the nuclear envelope by an inner nuclear membrane protein Bqt4 in fission yeast.
Nucleic Acids Res., 47, 2019
|
|
4ZMI
 
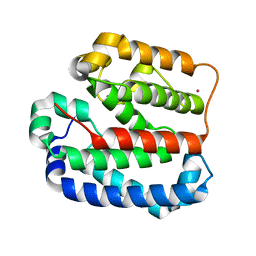 | | Crystal structure of the Helical domain of S. pombe Taz1 | | Descriptor: | COBALT (II) ION, MAGNESIUM ION, Telomere length regulator taz1 | | Authors: | Deng, W, Wu, J, Wang, F, Lei, M. | | Deposit date: | 2015-05-04 | | Release date: | 2015-09-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Fission yeast telomere-binding protein Taz1 is a functional but not a structural counterpart of human TRF1 and TRF2.
Cell Res., 25, 2015
|
|
6A6W
 
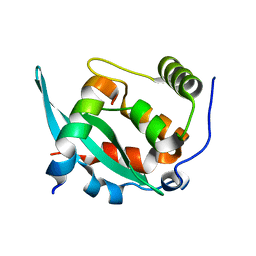 | |
4ZMK
 
 | |
1WLI
 
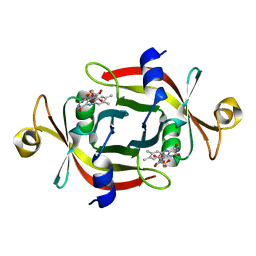 | |
2D1J
 
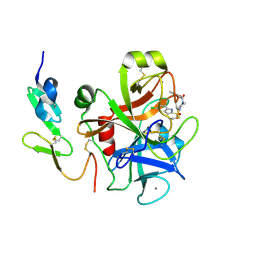 | | Factor Xa in complex with the inhibitor 2-[[4-[(5-chloroindol-2-yl)sulfonyl]piperazin-1-yl] carbonyl]thieno[3,2-b]pyridine n-oxide | | Descriptor: | 2-({4-[(5-CHLORO-1H-INDOL-2-YL)SULFONYL]PIPERAZIN-1-YL}CARBONYL)THIENO[3,2-B]PYRIDINE 4-OXIDE, CALCIUM ION, Coagulation factor X, ... | | Authors: | Suzuki, M. | | Deposit date: | 2005-08-24 | | Release date: | 2006-08-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Design, synthesis, and biological activity of non-basic compounds as factor Xa inhibitors: SAR study of S1 and aryl binding sites.
Bioorg.Med.Chem., 13, 2005
|
|
3A20
 
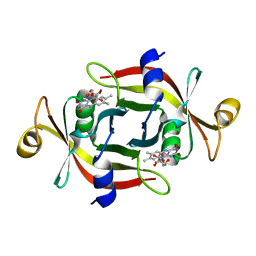 | |
3QEE
 
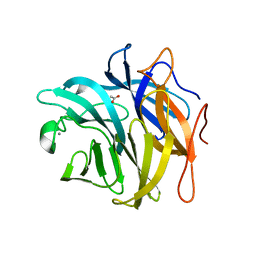 | | The structure and function of an arabinan-specific alpha-1,2-arabinofuranosidase identified from screening the activities of bacterial GH43 glycoside hydrolases | | Descriptor: | ACETATE ION, Beta-xylosidase/alpha-L-arabinfuranosidase, gly43N, ... | | Authors: | Cartmell, A, Mckee, L.S, Pena, M, Larsbrink, J, Brumer, H, Lewis, R.J, Viks-Nielsen, A, Gilbert, H.J, Marles-Wright, J. | | Deposit date: | 2011-01-20 | | Release date: | 2011-02-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | The Structure and Function of an Arabinan-specific {alpha}-1,2-Arabinofuranosidase Identified from Screening the Activities of Bacterial GH43 Glycoside Hydrolases.
J.Biol.Chem., 286, 2011
|
|
3QED
 
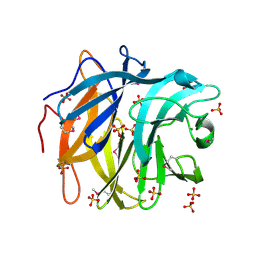 | | The structure and function of an arabinan-specific alpha-1,2-arabinofuranosidase identified from screening the activities of bacterial GH43 glycoside hydrolases | | Descriptor: | Beta-xylosidase/alpha-L-arabinfuranosidase, gly43N, CALCIUM ION, ... | | Authors: | Cartmell, A, Mckee, L.S, Pena, M, Larsbrink, J, Brumer, H, Lewis, R.J, Viks-Nielsen, A, Gilbert, H.J, Marles-Wright, J. | | Deposit date: | 2011-01-20 | | Release date: | 2011-02-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | The Structure and Function of an Arabinan-specific {alpha}-1,2-Arabinofuranosidase Identified from Screening the Activities of Bacterial GH43 Glycoside Hydrolases.
J.Biol.Chem., 286, 2011
|
|
3A6R
 
 | | E13Q mutant of FMN-binding protein from Desulfovibrio vulgaris (Miyazaki F) | | Descriptor: | CHLORIDE ION, FLAVIN MONONUCLEOTIDE, FMN-binding protein | | Authors: | Nakanishi, T, Haruyama, Y, Inoue, H, Kitamura, M. | | Deposit date: | 2009-09-08 | | Release date: | 2010-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Effects of the disappearance of one charge on ultrafast fluorescence dynamics of the FMN binding protein.
J.Phys.Chem.B, 114, 2010
|
|
3A6Q
 
 | | E13T mutant of FMN-binding protein from Desulfovibrio vulgaris (Miyazaki F) | | Descriptor: | CHLORIDE ION, FLAVIN MONONUCLEOTIDE, FMN-binding protein | | Authors: | Nakanishi, T, Haruyama, Y, Inoue, H, Kitamura, M. | | Deposit date: | 2009-09-08 | | Release date: | 2010-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Effects of the disappearance of one charge on ultrafast fluorescence dynamics of the FMN binding protein.
J.Phys.Chem.B, 114, 2010
|
|
3QEF
 
 | | The structure and function of an arabinan-specific alpha-1,2-arabinofuranosidase identified from screening the activities of bacterial GH43 glycoside hydrolases | | Descriptor: | 1,2-ETHANEDIOL, Beta-xylosidase/alpha-L-arabinfuranosidase, gly43N, ... | | Authors: | Cartmell, A, Mckee, L.S, Pena, M, Larsbrink, J, Brumer, H, Lewis, R.J, Viks-Nielsen, A, Gilbert, H.J, Marles-Wright, J. | | Deposit date: | 2011-01-20 | | Release date: | 2011-02-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.789 Å) | | Cite: | The Structure and Function of an Arabinan-specific {alpha}-1,2-Arabinofuranosidase Identified from Screening the Activities of Bacterial GH43 Glycoside Hydrolases.
J.Biol.Chem., 286, 2011
|
|
3ANW
 
 | | A protein complex essential initiation of DNA replication | | Descriptor: | Putative uncharacterized protein | | Authors: | Oyama, T, Shirai, T, Nagasawa, N, Ishino, Y, Morikawa, K. | | Deposit date: | 2010-09-10 | | Release date: | 2011-07-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Architectures of archaeal GINS complexes, essential DNA replication initiation factors
Bmc Biol., 9, 2011
|
|
1JLG
 
 | | CRYSTAL STRUCTURE OF Y188C MUTANT HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH UC-781 | | Descriptor: | 2-METHYL-FURAN-3-CARBOTHIOIC ACID [4-CHLORO-3-(3-METHYL-BUT-2-ENYLOXY)-PHENYL]-AMIDE, HIV-1 RT A-chain, HIV-1 RT B-chain | | Authors: | Ren, J, Nichols, C, Bird, L, Chamberlain, P, Weaver, K, Short, S, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2001-07-16 | | Release date: | 2001-10-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural mechanisms of drug resistance for mutations at codons 181 and 188 in HIV-1 reverse transcriptase and the improved resilience of second generation non-nucleoside inhibitors.
J.Mol.Biol., 312, 2001
|
|
1JLB
 
 | | CRYSTAL STRUCTURE OF Y181C MUTANT HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH NEVIRAPINE | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, HIV-1 RT A-chain, HIV-1 RT B-chain | | Authors: | Ren, J, Nichols, C, Bird, L, Chamberlain, P, Weaver, K, Short, S, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2001-07-16 | | Release date: | 2001-10-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural mechanisms of drug resistance for mutations at codons 181 and 188 in HIV-1 reverse transcriptase and the improved resilience of second generation non-nucleoside inhibitors.
J.Mol.Biol., 312, 2001
|
|
1JLQ
 
 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH 739W94 | | Descriptor: | 2-AMINO-6-(3,5-DIMETHYLPHENYL)SULFONYLBENZONITRILE, HIV-1 RT, A-CHAIN, ... | | Authors: | Ren, J, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2001-07-16 | | Release date: | 2001-08-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | 2-Amino-6-arylsulfonylbenzonitriles as non-nucleoside reverse transcriptase inhibitors of HIV-1.
J.Med.Chem., 44, 2001
|
|
1JLC
 
 | | CRYSTAL STRUCTURE OF Y181C MUTANT HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH PETT-2 | | Descriptor: | HIV-1 RT A-chain, HIV-1 RT B-chain, N-[[3-FLUORO-4-ETHOXY-PYRID-2-YL]ETHYL]-N'-[5-CHLORO-PYRIDYL]-THIOUREA | | Authors: | Ren, J, Nichols, C, Bird, L, Chamberlain, P, Weaver, K, Short, S, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2001-07-16 | | Release date: | 2001-10-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural mechanisms of drug resistance for mutations at codons 181 and 188 in HIV-1 reverse transcriptase and the improved resilience of second generation non-nucleoside inhibitors.
J.Mol.Biol., 312, 2001
|
|
