6F58
 
 | | Crystal structure of human Brachyury (T) in complex with DNA | | Descriptor: | Brachyury protein, DNA (5'-D(*AP*AP*TP*TP*TP*CP*AP*CP*AP*CP*CP*TP*AP*GP*GP*TP*GP*TP*GP*AP*AP*AP*TP*T)-3'), SODIUM ION | | Authors: | Newman, J.A, Gavard, A.E, Krojer, T, Shrestha, L, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2017-12-01 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.253 Å) | | Cite: | Crystal structure of human Brachyury (T) in complex with DNA
To Be Published
|
|
6MR2
 
 | |
6MUJ
 
 | | Formylglycine generating enzyme bound to copper | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, CALCIUM ION, COPPER (II) ION, ... | | Authors: | Lafrance-Vanasse, J, Appel, M.J, Tsai, C.-L, Bertozzi, C, Tainer, J.A. | | Deposit date: | 2018-10-23 | | Release date: | 2019-02-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.249 Å) | | Cite: | Formylglycine-generating enzyme binds substrate directly at a mononuclear Cu(I) center to initiate O2activation.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6FD8
 
 | | Gamma-s crystallin dimer | | Descriptor: | Beta-crystallin S | | Authors: | Mabbitt, P.D, Thorn, D.C, Jackson, C.J, Carver, J.A. | | Deposit date: | 2017-12-22 | | Release date: | 2018-12-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Structure and Stability of the Disulfide-Linked gamma S-Crystallin Dimer Provide Insight into Oxidation Products Associated with Lens Cataract Formation.
J. Mol. Biol., 431, 2019
|
|
1PIW
 
 | | APO AND HOLO STRUCTURES OF AN NADP(H)-DEPENDENT CINNAMYL ALCOHOL DEHYDROGENASE FROM SACCHAROMYCES CEREVISIAE | | Descriptor: | Hypothetical zinc-type alcohol dehydrogenase-like protein in PRE5-FET4 intergenic region, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ZINC ION | | Authors: | Valencia, E, Larroy, C, Ochoa, W.F, Pares, X, Fita, I, Biosca, J.A. | | Deposit date: | 2003-05-30 | | Release date: | 2004-08-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Apo and Holo Structures of an NADP(H)-dependent Cinnamyl Alcohol Dehydrogenase from Saccharomyces cerevisiae
J.Mol.Biol., 341, 2004
|
|
6N4A
 
 | |
6OZY
 
 | | Wild type GapR crystal structure 2 from C. crescentus | | Descriptor: | CADMIUM ION, UPF0335 protein CC_3319 | | Authors: | Tarry, M, Harmel, C, Taylor, J.A, Marczynski, G.T, Schmeing, T.M. | | Deposit date: | 2019-05-16 | | Release date: | 2019-11-27 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.014 Å) | | Cite: | Structures of GapR reveal a central channel which could accommodate B-DNA.
Sci Rep, 9, 2019
|
|
6MHC
 
 | | Glutathione S-Transferase Omega 1 bound to covalent inhibitor 37 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Petrunak, E.M, Stuckey, J.A. | | Deposit date: | 2018-09-17 | | Release date: | 2019-02-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Design of N-(5-Phenylthiazol-2-yl)acrylamides as Novel and Potent Glutathione S-Transferase Omega 1 Inhibitors.
J. Med. Chem., 62, 2019
|
|
6MN7
 
 | | Cryo-EM structure of BG505.SOSIP.664 in complex with BF520.1 antigen binding fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BF520.1 Fab variable region, ... | | Authors: | Williams, J.A, Lee, K.K, Overbaugh, J. | | Deposit date: | 2018-10-01 | | Release date: | 2019-03-06 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Kappa chain maturation helps drive rapid development of an infant HIV-1 broadly neutralizing antibody lineage.
Nat Commun, 10, 2019
|
|
6MS3
 
 | | Crystal structure of the GH43 protein BlXynB mutant (K247S) from Bacillus licheniformis | | Descriptor: | CALCIUM ION, GLYCEROL, Glycoside Hydrolase Family 43, ... | | Authors: | Zanphorlin, L.M, Morais, M.A.B, Diogo, J.A, Murakami, M.T. | | Deposit date: | 2018-10-16 | | Release date: | 2019-04-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-guided design combined with evolutionary diversity led to the discovery of the xylose-releasing exo-xylanase activity in the glycoside hydrolase family 43.
Biotechnol. Bioeng., 116, 2019
|
|
6FC4
 
 | |
6FCF
 
 | | CHK1 KINASE IN COMPLEX WITH COMPOUND 44 | | Descriptor: | 4-[[(2~{R},3~{S})-2-methylpiperidin-3-yl]amino]-2-phenyl-thieno[3,2-c]pyridine-7-carboxamide, SULFATE ION, Serine/threonine-protein kinase Chk1 | | Authors: | Read, J.A, Breed, J. | | Deposit date: | 2017-12-20 | | Release date: | 2018-01-17 | | Last modified: | 2018-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Adventures in Scaffold Morphing: Discovery of Fused Ring Heterocyclic Checkpoint Kinase 1 (CHK1) Inhibitors.
J. Med. Chem., 61, 2018
|
|
6FCQ
 
 | |
6MXF
 
 | | MicroED structure of thiostrepton at 1.9 A resolution | | Descriptor: | Thiostrepton | | Authors: | Jones, C.G, Martynowycz, M.W, Hattne, J, Fulton, T, Stoltz, B.M, Rodriguez, J.A, Nelson, H.M, Gonen, T. | | Deposit date: | 2018-10-30 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-15 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.91 Å) | | Cite: | The CryoEM Method MicroED as a Powerful Tool for Small Molecule Structure Determination.
ACS Cent Sci, 4, 2018
|
|
6FFH
 
 | | Crystal Structure of mGluR5 in complex with Fenobam at 2.65 A | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-(3-chlorophenyl)-3-(3-methyl-5-oxidanylidene-4~{H}-imidazol-2-yl)urea, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Christopher, J.A, Orgovan, Z, Congreve, M, Dore, A.S, Errey, J.C, Marshall, F.H, Mason, J.S, Okrasa, K, Rucktooa, P, Serrano-Vega, M.J, Ferenczy, G.G, Keseru, G.M. | | Deposit date: | 2018-01-08 | | Release date: | 2018-03-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure-Based Optimization Strategies for G Protein-Coupled Receptor (GPCR) Allosteric Modulators: A Case Study from Analyses of New Metabotropic Glutamate Receptor 5 (mGlu5) X-ray Structures.
J.Med.Chem., 62, 2019
|
|
6MR6
 
 | |
6FCS
 
 | | The X-ray Structure of Lytic Transglycosylase Slt inactive mutant E503Q from Pseudomonas aeruginosa in complex with NAG-NAMpentapeptide-NAG-NAMpentapeptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-3-O-[(2R)-1-amino-1-oxopropan-2-yl]-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-methyl 2-acetamido-3-O-[(1R)-1-carboxyethyl]-2-deoxy-beta-D-glucopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-methyl 2-acetamido-2-deoxy-beta-D-glucopyranoside, ACETATE ION, ... | | Authors: | Batuecas, M.T, Dominguez-Gil, T, Hermoso, J.A. | | Deposit date: | 2017-12-21 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Exolytic and endolytic turnover of peptidoglycan by lytic transglycosylase Slt ofPseudomonas aeruginosa.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
1JCV
 
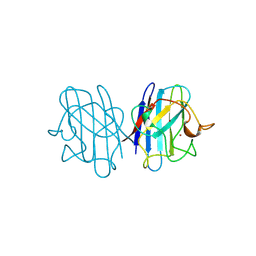 | | REDUCED BRIDGE-BROKEN YEAST CU/ZN SUPEROXIDE DISMUTASE LOW TEMPERATURE (-180C) STRUCTURE | | Descriptor: | COPPER (II) ION, CU/ZN SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Ogihara, N.L, Parge, H.E, Hart, P.J, Weiss, M.S, Valentine, J.S, Eisenberg, D.S, Tainer, J.A. | | Deposit date: | 1995-12-07 | | Release date: | 1996-03-08 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Unusual trigonal-planar copper configuration revealed in the atomic structure of yeast copper-zinc superoxide dismutase.
Biochemistry, 35, 1996
|
|
6N3Z
 
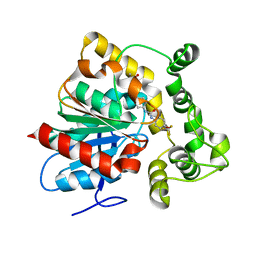 | | Crystal structure of an epoxide hydrolase from Trichoderma reesei in complex with inhibitor 4 | | Descriptor: | Epoxide hydrolase TrEH, N-methyl-4-{[trans-4-({[4-(trifluoromethoxy)phenyl]carbamoyl}amino)cyclohexyl]oxy}benzamide | | Authors: | de Oliveira, G.S, Adriani, P.P, Ribeiro, J.A, Morisseau, C, Hammock, B.D, Dias, M.V, Chambergo, F.S. | | Deposit date: | 2018-11-16 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.238 Å) | | Cite: | The molecular structure of an epoxide hydrolase from Trichoderma reesei in complex with urea or amide-based inhibitors.
Int. J. Biol. Macromol., 129, 2019
|
|
6PRY
 
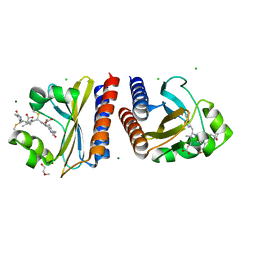 | | X-ray crystal structure of the blue-light absorbing state of PixJ from Thermosynechococcus elongatus by serial femtosecond crystallographic analysis | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Burgie, E.S, Clinger, J.A, Miller, M.D, Phillips Jr, G.N, Vierstra, R.D, Orville, A.M, Kern, J.F. | | Deposit date: | 2019-07-12 | | Release date: | 2019-12-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Photoreversible interconversion of a phytochrome photosensory module in the crystalline state.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6P7M
 
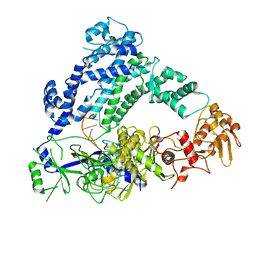 | | Cryo-EM structure of LbCas12a-crRNA: AcrVA4 (1:2 complex) | | Descriptor: | Cas12a, MAGNESIUM ION, anti-CRISPR VA4, ... | | Authors: | Knott, G.J, Liu, J.J, Doudna, J.A. | | Deposit date: | 2019-06-06 | | Release date: | 2019-08-21 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for AcrVA4 inhibition of specific CRISPR-Cas12a.
Elife, 8, 2019
|
|
6FBT
 
 | | The X-ray Structure of Lytic Transglycosylase Slt from Pseudomonas aeruginosa in complex with the reaction product NAG-anhNAMpentapeptide | | Descriptor: | 2-(2-ACETYLAMINO-4-HYDROXY-6,8-DIOXA-BICYCLO[3.2.1]OCT-3-YLOXY)-PROPIONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, NAG-anhNAMpentapeptide, ... | | Authors: | Batuecas, M.T, Dominguez-Gil, T, Hermoso, J.A. | | Deposit date: | 2017-12-19 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Exolytic and endolytic turnover of peptidoglycan by lytic transglycosylase Slt ofPseudomonas aeruginosa.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6N5H
 
 | | Crystal structure of an epoxide hydrolase from Trichoderma reesei in complex with inhibitor 5 | | Descriptor: | 4-[(trans-4-{[(3s,5s,7s)-tricyclo[3.3.1.1~3,7~]dec-1-ylcarbamoyl]amino}cyclohexyl)oxy]benzoic acid, Epoxide hydrolase TrEH | | Authors: | Oliveira, G.S, Adriano, P.P, Ribeiro, J.A, Morisseau, C, Hammock, B.D, Dias, M.V, Chambergo, F.S. | | Deposit date: | 2018-11-21 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.717 Å) | | Cite: | The molecular structure of an epoxide hydrolase from Trichoderma reesei in complex with urea or amide-based inhibitors
Int. J. Biol. Macromol., 129, 2019
|
|
6FC8
 
 | | CHK1 KINASE IN COMPLEX WITH COMPOUND 13 | | Descriptor: | 2-(3-fluorophenyl)-4-[[(3~{S})-piperidin-3-yl]amino]thieno[3,2-c]pyridine-7-carboxamide, SULFATE ION, Serine/threonine-protein kinase Chk1 | | Authors: | Read, J.A, Breed, J. | | Deposit date: | 2017-12-20 | | Release date: | 2018-01-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Adventures in Scaffold Morphing: Discovery of Fused Ring Heterocyclic Checkpoint Kinase 1 (CHK1) Inhibitors.
J. Med. Chem., 61, 2018
|
|
6FCK
 
 | | CHK1 KINASE IN COMPLEX WITH COMPOUND 13 | | Descriptor: | 2-phenyl-4-[[(3~{S})-piperidin-3-yl]amino]-1~{H}-indole-7-carboxamide, SULFATE ION, Serine/threonine-protein kinase Chk1 | | Authors: | Read, J.A, Breed, J. | | Deposit date: | 2017-12-20 | | Release date: | 2018-01-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Adventures in Scaffold Morphing: Discovery of Fused Ring Heterocyclic Checkpoint Kinase 1 (CHK1) Inhibitors.
J. Med. Chem., 61, 2018
|
|
