7QA9
 
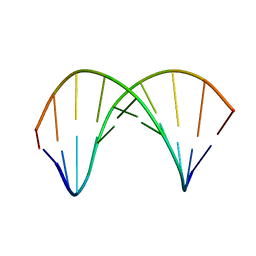 | | 10bp DNA/DNA duplex | | Descriptor: | DNA (5'-D(*CP*CP*AP*TP*TP*AP*TP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*AP*TP*AP*AP*TP*GP*G)-3') | | Authors: | Li, Q, Trajkovski, M, Fan, C, Chen, J, Zhou, Y, Lu, K, Li, H, Su, X, Xi, Z, Plavec, J, Zhou, C. | | Deposit date: | 2021-11-16 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | 4'-SCF 3 -Labeling Constitutes a Sensitive 19 F NMR Probe for Characterization of Interactions in the Minor Groove of DNA.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
6LUK
 
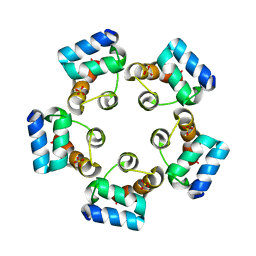 | |
6LUJ
 
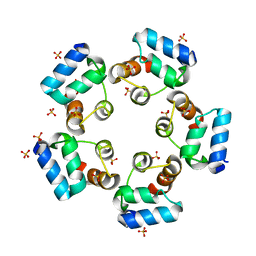 | | Crystal structure of the SAMD1 SAM domain | | Descriptor: | Atherin, SULFATE ION | | Authors: | Cao, Y, Zhou, Y, Wang, Z. | | Deposit date: | 2020-01-29 | | Release date: | 2021-02-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | The SAM domain-containing protein 1 (SAMD1) acts as a repressive chromatin regulator at unmethylated CpG islands.
Sci Adv, 7, 2021
|
|
6L17
 
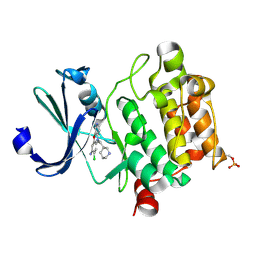 | | Crystal structure of Ser/Thr kinase Pim1 in complex with 10-DEBC derivatives | | Descriptor: | 7-chloranyl-5-[3-[(3~{S})-piperidin-3-yl]propyl]pyrido[3,4-b][1,4]benzoxazin-8-amine, Serine/threonine-protein kinase pim-1 | | Authors: | Zhang, W, Xie, Y, Cao, R, Huang, N, Zhou, Y. | | Deposit date: | 2019-09-27 | | Release date: | 2020-09-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure-Based Optimization of 10-DEBC Derivatives as Potent and Selective Pim-1 Kinase Inhibitors.
J.Chem.Inf.Model., 60, 2020
|
|
6L11
 
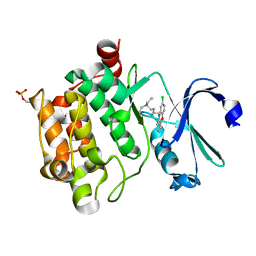 | | Crystal structure of Ser/Thr kinase Pim1 in complex with 10-DEBC derivatives | | Descriptor: | 5-(2-chloranylphenoxazin-10-yl)-~{N},~{N}-diethyl-pentan-1-amine, Serine/threonine-protein kinase pim-1 | | Authors: | Zhang, W, Xie, Y, Cao, R, Huang, N, Zhou, Y. | | Deposit date: | 2019-09-27 | | Release date: | 2020-05-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure-Based Optimization of 10-DEBC Derivatives as Potent and Selective Pim-1 Kinase Inhibitors.
J.Chem.Inf.Model., 60, 2020
|
|
6L13
 
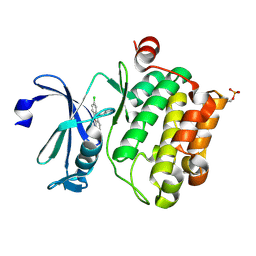 | | Crystal structure of Ser/Thr kinase Pim1 in complex with 10-DEBC derivatives | | Descriptor: | 2-chloranyl-10-(2-piperidin-4-ylethyl)phenoxazine, Serine/threonine-protein kinase pim-1 | | Authors: | Zhang, W, Xie, Y, Cao, R, Huang, N, Zhou, Y. | | Deposit date: | 2019-09-27 | | Release date: | 2020-05-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structure-Based Optimization of 10-DEBC Derivatives as Potent and Selective Pim-1 Kinase Inhibitors.
J.Chem.Inf.Model., 60, 2020
|
|
6L14
 
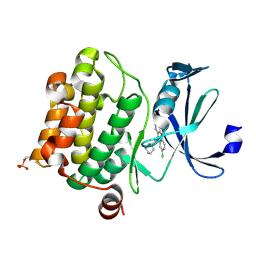 | | Crystal structure of Ser/Thr kinase Pim1 in complex with 10-DEBC derivatives | | Descriptor: | 2-chloranyl-10-[3-[(3~{S})-piperidin-3-yl]propyl]phenoxazine, Serine/threonine-protein kinase pim-1 | | Authors: | Zhang, W, Xie, Y, Cao, R, Huang, N, Zhou, Y. | | Deposit date: | 2019-09-27 | | Release date: | 2020-05-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-Based Optimization of 10-DEBC Derivatives as Potent and Selective Pim-1 Kinase Inhibitors.
J.Chem.Inf.Model., 60, 2020
|
|
2DGN
 
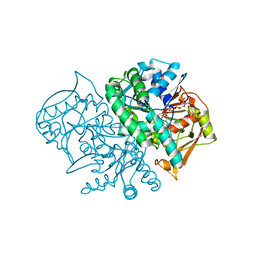 | | Mouse Muscle Adenylosuccinate Synthetase partially ligated complex with GTP, 2'-deoxy-IMP | | Descriptor: | 9-(2-DEOXY-5-O-PHOSPHONO-BETA-D-ERYTHRO-PENTOFURANOSYL)-6-(PHOSPHONOOXY)-9H-PURINE, Adenylosuccinate synthetase isozyme 1, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Iancu, C.V, Zhou, Y, Borza, T, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2006-03-15 | | Release date: | 2006-09-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Cavitation as a mechanism of substrate discrimination by adenylosuccinate synthetases.
Biochemistry, 45, 2006
|
|
2AY0
 
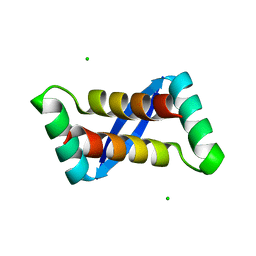 | | Structure of the Lys9Met mutant of the E. coli Proline Utilization A (PutA) DNA-binding domain. | | Descriptor: | Bifunctional putA protein, CHLORIDE ION | | Authors: | Larson, J.D, Schuermann, J.P, Zhou, Y, Jenkins, J.L, Becker, D.F, Tanner, J.J. | | Deposit date: | 2005-09-06 | | Release date: | 2006-08-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of the DNA-binding domain of Escherichia coli proline utilization A flavoprotein and analysis of the role of Lys9 in DNA recognition.
Protein Sci., 15, 2006
|
|
3O0J
 
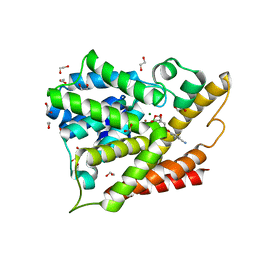 | | PDE4B In complex with ligand an2898 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(1-hydroxy-1,3-dihydro-2,1-benzoxaborol-5-yl)oxy]benzene-1,2-dicarbonitrile, MAGNESIUM ION, ... | | Authors: | Alley, M.R.K, Zhou, Y. | | Deposit date: | 2010-07-19 | | Release date: | 2011-08-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Boron-based phosphodiesterase inhibitors show novel binding of boron to PDE4 bimetal center.
Febs Lett., 586, 2012
|
|
6NJD
 
 | |
6NII
 
 | |
6LDU
 
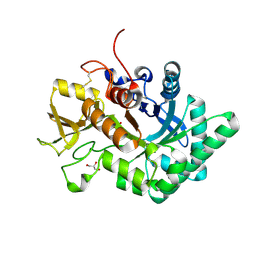 | |
6LE7
 
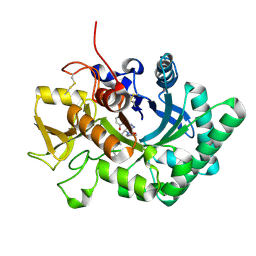 | | Crystal structure of nematode family I chitinase,CeCht1, in complex with dihydropyrrolopyrazol-6-one derivate 2 | | Descriptor: | (4R)-3-(2-hydroxyphenyl)-4-(3-methoxy-4-propoxy-phenyl)-5-(pyridin-3-ylmethyl)-1,4-dihydropyrrolo[3,4-c]pyrazol-6-one, Probable endochitinase | | Authors: | Chen, Q, Yang, Q, Zhou, Y. | | Deposit date: | 2019-11-24 | | Release date: | 2021-05-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85753441 Å) | | Cite: | Crystal structure of nematode family I chitinase,CeCht1, in complex with dihydropyrrolopyrazol-6-one derivate 2
To Be Published
|
|
6LE8
 
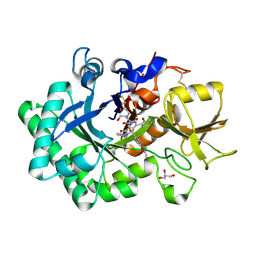 | | Crystal structure of nematode family I chitinase,CeCht1, in complex with dihydropyrrolopyrazol-6-one derivate 1 | | Descriptor: | (4R)-4-(4-ethoxyphenyl)-3-(2-hydroxyphenyl)-5-(pyridin-3-ylmethyl)-1,4-dihydropyrrolo[3,4-c]pyrazol-6-one, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, ... | | Authors: | Chen, Q, Yang, Q, Zhou, Y. | | Deposit date: | 2019-11-24 | | Release date: | 2021-05-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.39909327 Å) | | Cite: | Crystal structure of nematode family I chitinase,CeCht1, in complex with dihydropyrrolopyrazol-6-one derivate 1
To Be Published
|
|
6MDR
 
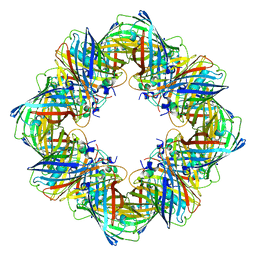 | | Cryo-EM structure of the Ceru+32/GFP-17 protomer | | Descriptor: | Ceru+32, GFP-17 | | Authors: | Simon, A.J, Zhou, Y, Ramasubramani, V, Glaser, J, Pothukuchy, A, Golihar, J, Gerberich, J.C, Leggere, J.C, Morrow, B.R, Jung, C, Glotzer, S.C, Taylor, D.W, Ellington, A.D. | | Deposit date: | 2018-09-05 | | Release date: | 2019-01-23 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Supercharging enables organized assembly of synthetic biomolecules.
Nat Chem, 11, 2019
|
|
7TAD
 
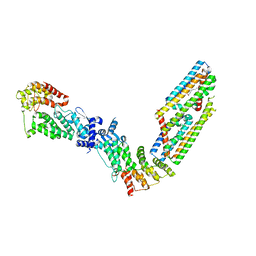 | | CryoEM structure of the (NPR1)2-(TGA3)2 complex | | Descriptor: | PALMITIC ACID, Regulatory protein NPR1, Transcription factor TGA3, ... | | Authors: | Wu, Q, Zhou, Y, Bartesaghi, A, Dong, X, Zhou, P. | | Deposit date: | 2021-12-20 | | Release date: | 2022-03-16 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of NPR1 in activating plant immunity.
Nature, 605, 2022
|
|
7TAC
 
 | | Cryo-EM structure of the (TGA3)2-(NPR1)2-(TGA3)2 complex | | Descriptor: | PALMITIC ACID, Regulatory protein NPR1, Transcription factor TGA3, ... | | Authors: | Wu, Q, Zhou, Y, Bartesaghi, A, Dong, X, Zhou, P. | | Deposit date: | 2021-12-20 | | Release date: | 2022-03-16 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of NPR1 in activating plant immunity.
Nature, 605, 2022
|
|
3WNV
 
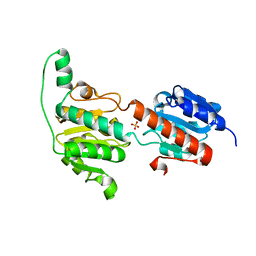 | | Crystal structure of a glyoxylate reductase from Paecilomyes thermophila | | Descriptor: | SULFATE ION, glyoxylate reductase | | Authors: | Duan, X, Hu, S, Zhou, P, Zhou, Y, Jiang, Z. | | Deposit date: | 2013-12-17 | | Release date: | 2014-12-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Characterization and crystal structure of a first fungal glyoxylate reductase from Paecilomyes thermophila
Enzyme.Microb.Technol., 60, 2014
|
|
6WZT
 
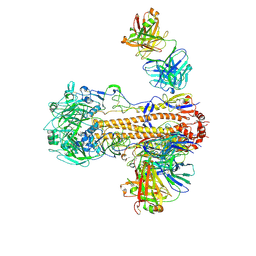 | | CryoEM structure of influenza hemagglutinin A/Victoria/361/2011 in complex with cyno antibody 3B10 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cyno antibody heavy chain, ... | | Authors: | Qiu, Y, Zhou, Y, Darricarrere, N. | | Deposit date: | 2020-05-14 | | Release date: | 2021-03-10 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Broad neutralization of H1 and H3 viruses by adjuvanted influenza HA stem vaccines in nonhuman primates.
Sci Transl Med, 13, 2021
|
|
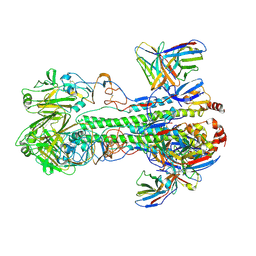
6XGC
 
 | |
6PDX
 
 | |
6XDG
 
 | | Complex of SARS-CoV-2 receptor binding domain with the Fab fragments of two neutralizing antibodies | | Descriptor: | REGN10933 antibody Fab fragment heavy chain, REGN10933 antibody Fab fragment light chain, REGN10987 antibody Fab fragment heavy chain, ... | | Authors: | Franklin, M.C, Saotome, K, Romero Hernandez, A, Zhou, Y. | | Deposit date: | 2020-06-10 | | Release date: | 2020-06-24 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail.
Science, 369, 2020
|
|
8DTI
 
 | | Cryo-EM structure of Arabidopsis SPY in complex with GDP-fucose | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE-BETA-L-FUCOPYRANOSE, Probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SPINDLY | | Authors: | Kumar, S, Zhou, Y, Dillard, L, Borgnia, M.J, Bartesaghi, A, Zhou, P. | | Deposit date: | 2022-07-25 | | Release date: | 2023-03-08 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structure of the full length Arabidopsis SPY with complete TPRs
Nat Commun, 2023
|
|
8DTG
 
 | | Cryo-EM structure of Arabidopsis SPY alternative conformation 1 | | Descriptor: | Probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SPINDLY | | Authors: | Kumar, S, Zhou, Y, Dillard, L, Borgnia, M.J, Bartesaghi, A, Zhou, P. | | Deposit date: | 2022-07-25 | | Release date: | 2023-03-08 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structure of the full length Arabidopsis SPY with complete TPRs
Nat Commun, 2023
|
|
