2YK7
 
 | | Structure of Neisseria LOS-specific sialyltransferase (NST), in complex with CMP-3F-Neu5Ac. | | Descriptor: | 1,2-ETHANEDIOL, CMP-N-ACETYLNEURAMINATE-BETA-GALACTOSAMIDE-ALPHA-2,3-SIALYLTRANSFERASE, CYTIDINE-5'-MONOPHOSPHATE-3-FLUORO-N-ACETYL-NEURAMINIC ACID, ... | | Authors: | Lin, L.Y.C, Rakic, B, Chiu, C.P.C, Lameignere, E, Wakarchuk, W.W, Withers, S.G, Strynadka, N.C.J. | | Deposit date: | 2011-05-25 | | Release date: | 2011-08-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structure and Mechanism of the Lipooligosaccharide Sialyltransferase from Neisseria Meningitidis
J.Biol.Chem., 286, 2011
|
|
2YK4
 
 | | Structure of Neisseria LOS-specific sialyltransferase (NST). | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-{2-[2-(2-{2-[2-(2-{2-[4-(1,1,3,3-TETRAMETHYL-BUTYL)-PHENOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOX Y}-ETHOXY)-ETHANOL, CMP-N-ACETYLNEURAMINATE-BETA-GALACTOSAMIDE-ALPHA-2,3-SIALYLTRANSFERASE, ... | | Authors: | Lin, L.Y.C, Rakic, B, Chiu, C.P.C, Lameignere, E, Wakarchuk, W.W, Withers, S.G, Strynadka, N.C.J. | | Deposit date: | 2011-05-25 | | Release date: | 2011-08-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structure and Mechanism of the Lipooligosaccharide Sialyltransferase from Neisseria Meningitidis
J.Biol.Chem., 286, 2011
|
|
2YK5
 
 | | Structure of Neisseria LOS-specific sialyltransferase (NST), in complex with CMP. | | Descriptor: | 1,2-ETHANEDIOL, CMP-N-ACETYLNEURAMINATE-BETA-GALACTOSAMIDE-ALPHA-2,3-SIALYLTRANSFERASE, CYTIDINE-5'-MONOPHOSPHATE, ... | | Authors: | Lin, L.Y.C, Rakic, B, Chiu, C.P.C, Lameignere, E, Wakarchuk, W.W, Withers, S.G, Strynadka, N.C.J. | | Deposit date: | 2011-05-25 | | Release date: | 2011-08-31 | | Last modified: | 2019-09-25 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structure and Mechanism of the Lipooligosaccharide Sialyltransferase from Neisseria Meningitidis
J.Biol.Chem., 286, 2011
|
|
2YK6
 
 | | Structure of Neisseria LOS-specific sialyltransferase (NST), in complex with CDP. | | Descriptor: | CMP-N-ACETYLNEURAMINATE-BETA-GALACTOSAMIDE-ALPHA-2,3-SIALYLTRANSFERASE, CYTIDINE-5'-DIPHOSPHATE, PENTAETHYLENE GLYCOL, ... | | Authors: | Lin, L.Y.C, Rakic, B, Chiu, C.P.C, Lameignere, E, Wakarchuk, W.W, Withers, S.G, Strynadka, N.C.J. | | Deposit date: | 2011-05-25 | | Release date: | 2011-08-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Structure and Mechanism of the Lipooligosaccharide Sialyltransferase from Neisseria Meningitidis
J.Biol.Chem., 286, 2011
|
|
1AVS
 
 | |
3SPU
 
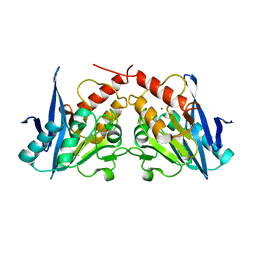 | | apo NDM-1 Crystal Structure | | Descriptor: | Beta-lactamase NDM-1, ZINC ION | | Authors: | Strynadka, N.C.J, King, D.T. | | Deposit date: | 2011-07-03 | | Release date: | 2011-08-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of New Delhi metallo-beta-lactamase reveals molecular basis for antibiotic resistance
Protein Sci., 20, 2011
|
|
3L7J
 
 | |
3L7K
 
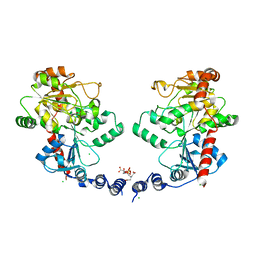 | | Structure of the Wall Teichoic Acid Polymerase TagF, H444N + CDPG (15 minute soak) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, SULFATE ION, ... | | Authors: | Strynadka, N.C.J, Lovering, A.L. | | Deposit date: | 2009-12-28 | | Release date: | 2010-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the bacterial teichoic acid polymerase TagF provides insights into membrane association and catalysis.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3L7I
 
 | |
1JTG
 
 | | CRYSTAL STRUCTURE OF TEM-1 BETA-LACTAMASE / BETA-LACTAMASE INHIBITOR PROTEIN COMPLEX | | Descriptor: | BETA-LACTAMASE INHIBITORY PROTEIN, BETA-LACTAMASE TEM, CALCIUM ION | | Authors: | Strynadka, N.C.J, Jensen, S.E, Alzari, P.M, James, M.N. | | Deposit date: | 2001-08-20 | | Release date: | 2001-10-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal structure and kinetic analysis of beta-lactamase inhibitor protein-II in complex with TEM-1 beta-lactamase.
Nat.Struct.Biol., 8, 2001
|
|
3GMU
 
 | | Crystal Structure of Beta-Lactamse Inhibitory Protein (BLIP) in Apo Form | | Descriptor: | AMMONIUM ION, Beta-lactamase inhibitory protein, SULFATE ION | | Authors: | Strynadka, N.C.J, Gretes, M, James, M.N.G. | | Deposit date: | 2009-03-15 | | Release date: | 2009-03-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Insights into positive and negative requirements for protein-protein interactions by crystallographic analysis of the beta-lactamase inhibitory proteins BLIP, BLIP-I, and BLP.
J.Mol.Biol., 389, 2009
|
|
1PPK
 
 | |
4EYB
 
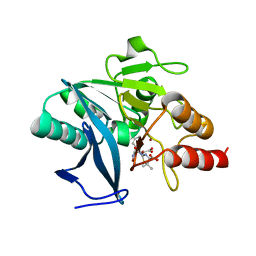 | | Crystal structure of NDM-1 bound to hydrolyzed oxacillin | | Descriptor: | (2R,4S)-2-[(R)-carboxy{[(5-methyl-3-phenyl-1,2-oxazol-4-yl)carbonyl]amino}methyl]-5,5-dimethyl-1,3-thiazolidine-4-carbo xylic acid, Beta-lactamase NDM-1, ZINC ION | | Authors: | Strynadka, N.C.J, King, D.T. | | Deposit date: | 2012-05-01 | | Release date: | 2012-08-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | New Delhi Metallo-Beta-Lactamase: Structural Insights into Beta-Lactam Recognition and Inhibition
J.Am.Chem.Soc., 134, 2012
|
|
4EY2
 
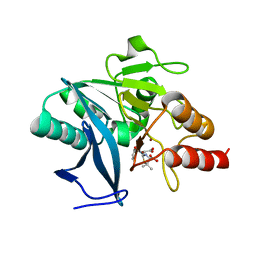 | | Crystal structure of NDM-1 bound to hydrolyzed methicillin | | Descriptor: | (2R,4S)-2-{(R)-carboxy[(2,6-dimethoxybenzoyl)amino]methyl}-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, Beta-lactamase NDM-1, ZINC ION | | Authors: | Strynadka, N.C.J, King, D.T. | | Deposit date: | 2012-05-01 | | Release date: | 2012-08-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | New Delhi Metallo-Beta-Lactamase: Structural Insights into Beta-Lactam Recognition and Inhibition
J.Am.Chem.Soc., 134, 2012
|
|
4EXY
 
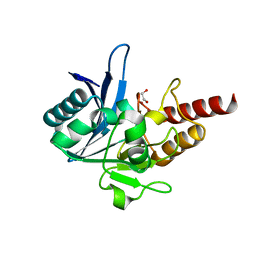 | |
4EYF
 
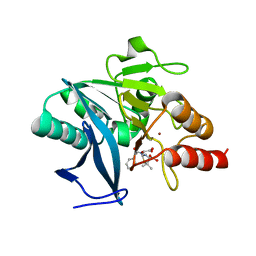 | | Crystal structure of NDM-1 bound to hydrolyzed benzylpenicillin | | Descriptor: | (2R,4S)-2-{(R)-carboxy[(phenylacetyl)amino]methyl}-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, Beta-lactamase NDM-1, ZINC ION | | Authors: | Strynadka, N.C.J, King, D.T. | | Deposit date: | 2012-05-01 | | Release date: | 2012-08-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | New Delhi Metallo-Beta-Lactamase: Structural Insights into Beta-Lactam Recognition and Inhibition
J.Am.Chem.Soc., 134, 2012
|
|
4EXS
 
 | | Crystal structure of NDM-1 bound to L-captopril | | Descriptor: | Beta-lactamase NDM-1, L-CAPTOPRIL, ZINC ION | | Authors: | Strynadka, N.C.J, King, D.T. | | Deposit date: | 2012-04-30 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | New Delhi Metallo-Beta-Lactamase: Structural Insights into Beta-Lactam Recognition and Inhibition
J.Am.Chem.Soc., 134, 2012
|
|
4EYL
 
 | | Crystal structure of NDM-1 bound to hydrolyzed meropenem | | Descriptor: | (2S)-2-[(1S,2R)-1-carboxy-2-hydroxypropyl]-4-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-3-methyl-2H-pyrro le-5-carboxylic acid, Beta-lactamase NDM-1, ZINC ION | | Authors: | Strynadka, N.C.J, King, D.T. | | Deposit date: | 2012-05-01 | | Release date: | 2012-08-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | New Delhi Metallo-Beta-Lactamase: Structural Insights into Beta-Lactam Recognition and Inhibition
J.Am.Chem.Soc., 134, 2012
|
|
3GMV
 
 | | Crystal Structure of Beta-Lactamse Inhibitory Protein-I (BLIP-I) in Apo Form | | Descriptor: | Beta-lactamase inhibitory protein BLIP-I, TRIS(HYDROXYETHYL)AMINOMETHANE | | Authors: | Lim, D.C, Gretes, M, Strynadka, N.C.J. | | Deposit date: | 2009-03-15 | | Release date: | 2009-03-31 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Insights into positive and negative requirements for protein-protein interactions by crystallographic analysis of the beta-lactamase inhibitory proteins BLIP, BLIP-I, and BLP.
J.Mol.Biol., 389, 2009
|
|
4KPG
 
 | | Crystal structure of MycP1 from the ESX-1 type VII secretion system | | Descriptor: | Membrane-anchored mycosin mycp1 | | Authors: | Solomonson, M, Wasney, G.A, Watanabe, N, Gruninger, R.J, Prehna, G, Strynadka, N.C.J. | | Deposit date: | 2013-05-13 | | Release date: | 2013-05-22 | | Last modified: | 2013-07-03 | | Method: | X-RAY DIFFRACTION (2.148 Å) | | Cite: | Structure of the Mycosin-1 Protease from the Mycobacterial ESX-1 Protein Type VII Secretion System.
J.Biol.Chem., 288, 2013
|
|
3UJZ
 
 | |
4OYC
 
 | |
2JW1
 
 | | Structural characterization of the type III pilotin-secretin interaction in Shigella flexneri by NMR spectroscopy | | Descriptor: | Lipoprotein mxiM, Outer membrane protein mxiD | | Authors: | Okon, M.S, Lario, P.I, Creagh, L, Jung, Y.M.T, Maurelli, A.T, Strynadka, N.C.J, McIntosh, L.P. | | Deposit date: | 2007-10-02 | | Release date: | 2008-09-02 | | Last modified: | 2020-02-19 | | Method: | SOLUTION NMR | | Cite: | Structural Characterization of the Type-III Pilot-Secretin Complex from Shigella flexneri
Structure, 16, 2008
|
|
4W4M
 
 | | Crystal structure of PrgK 19-92 | | Descriptor: | Lipoprotein PrgK | | Authors: | Bergeron, J.R.C, Strynadka, N.C.J. | | Deposit date: | 2014-08-15 | | Release date: | 2014-10-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The Modular Structure of the Inner-Membrane Ring Component PrgK Facilitates Assembly of the Type III Secretion System Basal Body.
Structure, 23, 2015
|
|
5CUL
 
 | | crystal structure of the PscU C-terminal domain | | Descriptor: | Translocation protein in type III secretion | | Authors: | Bergeron, J.R.C, Strynadka, N.C.J. | | Deposit date: | 2015-07-24 | | Release date: | 2015-12-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Structure of a Type 3 Secretion System (T3SS) Ruler Protein Suggests a Molecular Mechanism for Needle Length Sensing.
J.Biol.Chem., 291, 2016
|
|
