8OUH
 
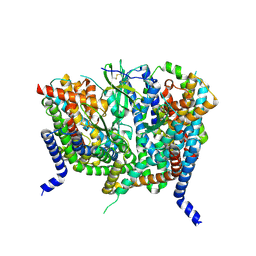 | | Complex of human ASCT2 with Syncytin-1 | | Descriptor: | ALANINE, Neutral amino acid transporter B(0), Syncytin-1 | | Authors: | Khare, S, Reyes, N. | | Deposit date: | 2023-04-23 | | Release date: | 2024-05-01 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Receptor-recognition and antiviral mechanisms of retrovirus-derived human proteins.
Nat.Struct.Mol.Biol., 31, 2024
|
|
4M3K
 
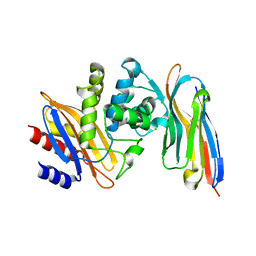 | | Structure of a single domain camelid antibody fragment cAb-H7S in complex with the BlaP beta-lactamase from Bacillus licheniformis | | Descriptor: | Beta-lactamase, CHLORIDE ION, Camelid heavy-chain antibody variable fragment cAb-H7S | | Authors: | Pain, C, Kerff, F, Herman, R, Sauvage, E, Preumont, S, Charlier, P, Dumoulin, M. | | Deposit date: | 2013-08-06 | | Release date: | 2014-08-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Probing the mechanism of aggregation of polyQ model proteins with camelid heavy-chain antibody fragments
To be Published
|
|
4M3J
 
 | | Structure of a single-domain camelid antibody fragment cAb-H7S specific of the BlaP beta-lactamase from Bacillus licheniformis | | Descriptor: | Camelid heavy-chain antibody variable fragment cAb-H7S, SULFATE ION | | Authors: | Pain, C, Kerff, F, Herman, R, Sauvage, E, Preumont, S, Charlier, P, Dumoulin, M. | | Deposit date: | 2013-08-06 | | Release date: | 2014-08-06 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Probing the mechanism of aggregation of polyQ model proteins with camelid heavy-chain antibody fragments
To be Published
|
|
9ENA
 
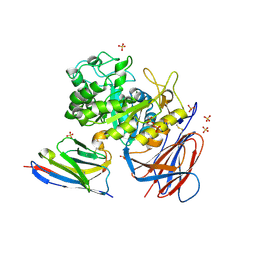 | | Lysosomal glucocerebrosidase in complex with a stabilizing nanobody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chains: B, ... | | Authors: | Dal Maso, T, Versees, W. | | Deposit date: | 2024-03-12 | | Release date: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Developing nanobodies as allosteric molecular chaperones of glucocerebrosidase function.
Nat Commun, 16, 2025
|
|
4N1H
 
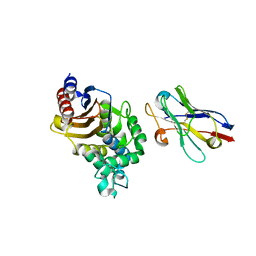 | | Structure of a single-domain camelid antibody fragment cAb-F11N in complex with the BlaP beta-lactamase from Bacillus licheniformis | | Descriptor: | Beta-lactamase, Camelid heavy-chain antibody variable fragment cAb-F11N | | Authors: | Pain, C, Kerff, F, Herman, R, Sauvage, E, Preumont, S, Charlier, P, Dumoulin, M. | | Deposit date: | 2013-10-04 | | Release date: | 2014-10-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Probing the mechanism of aggregation of polyQ model proteins with camelid heavy-chain antibody fragments
To be Published
|
|
4DK3
 
 | | Structure of Editosome protein | | Descriptor: | RNA-editing complex protein MP81, single domain antibody VHH | | Authors: | Park, Y.-J, Hol, W. | | Deposit date: | 2012-02-03 | | Release date: | 2012-07-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | The structure of the C-terminal domain of the largest editosome interaction protein and its role in promoting RNA binding by RNA-editing ligase L2.
Nucleic Acids Res., 40, 2012
|
|
4DKA
 
 | | Structure of Editosome protein | | Descriptor: | RNA-editing complex protein MP81, SODIUM ION, single domain antibody VHH | | Authors: | Park, Y.-J, Hol, W. | | Deposit date: | 2012-02-03 | | Release date: | 2012-07-04 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | The structure of the C-terminal domain of the largest editosome interaction protein and its role in promoting RNA binding by RNA-editing ligase L2.
Nucleic Acids Res., 40, 2012
|
|
4DK6
 
 | | Structure of Editosome protein | | Descriptor: | RNA-editing complex protein MP81, single domain antibody VHH | | Authors: | Park, Y.-J, Hol, W. | | Deposit date: | 2012-02-03 | | Release date: | 2012-07-04 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The structure of the C-terminal domain of the largest editosome interaction protein and its role in promoting RNA binding by RNA-editing ligase L2.
Nucleic Acids Res., 40, 2012
|
|
6ZG3
 
 | | the structure of ECF PanT transporter in a complex with a nanobody | | Descriptor: | CA14381 nanobody, CITRIC ACID, Conserved hypothetical membrane protein, ... | | Authors: | Setyawati, I, Guskov, A, Slotboom, D.J. | | Deposit date: | 2020-06-18 | | Release date: | 2021-03-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | In vitro reconstitution of dynamically interacting integral membrane subunits of energy-coupling factor transporters.
Elife, 9, 2020
|
|
6ZE1
 
 | | human NBD1 of CFTR in complex with nanobody G11a | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator, ... | | Authors: | Garcia-Pino, A, Govaerts, C, Scholl, D, Sigoillot, M. | | Deposit date: | 2020-06-15 | | Release date: | 2021-06-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.705 Å) | | Cite: | A topological switch in CFTR modulates channel activity and sensitivity to unfolding.
Nat.Chem.Biol., 17, 2021
|
|
7APJ
 
 | | Structure of autoinhibited Akt1 reveals mechanism of PIP3-mediated activation | | Descriptor: | NB41, RAC-alpha serine/threonine-protein kinase,Non-specific serine/threonine protein kinase,RAC-alpha serine/threonine-protein kinase | | Authors: | Truebestein, L, Hornegger, H, Leonard, T.A. | | Deposit date: | 2020-10-16 | | Release date: | 2021-08-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of autoinhibited Akt1 reveals mechanism of PIP 3 -mediated activation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6GS1
 
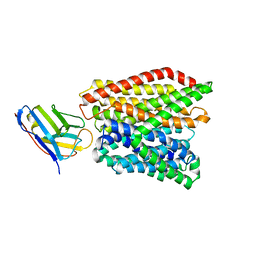 | | Crystal structure of peptide transporter DtpA-nanobody in MES buffer | | Descriptor: | Dipeptide and tripeptide permease A, Nanobody 00 | | Authors: | Ural-Blimke, Y, Flayhan, A, Loew, C, Quistgaard, E.M. | | Deposit date: | 2018-06-13 | | Release date: | 2019-01-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Structure of Prototypic Peptide Transporter DtpA from E. coli in Complex with Valganciclovir Provides Insights into Drug Binding of Human PepT1.
J. Am. Chem. Soc., 141, 2019
|
|
6GS4
 
 | | Crystal structure of peptide transporter DtpA-nanobody in complex with valganciclovir | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Dipeptide and tripeptide permease A, [(2~{S})-2-[(2-azanyl-6-oxidanylidene-3~{H}-purin-9-yl)methoxy]-3-oxidanyl-propyl] (2~{S})-2-azanyl-3-methyl-butanoate, ... | | Authors: | Ural-Blimke, Y, Flayhan, A, Quistgaard, E.M, Loew, C. | | Deposit date: | 2018-06-13 | | Release date: | 2019-01-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.645 Å) | | Cite: | Structure of Prototypic Peptide Transporter DtpA from E. coli in Complex with Valganciclovir Provides Insights into Drug Binding of Human PepT1.
J. Am. Chem. Soc., 141, 2019
|
|
6GS7
 
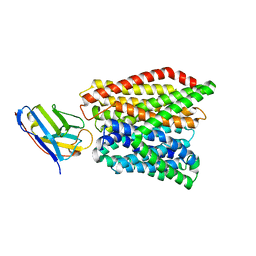 | | Crystal structure of peptide transporter DtpA-nanobody in glycine buffer | | Descriptor: | Dipeptide and tripeptide permease A, nanobody | | Authors: | Ural-Blimke, Y, Flayhan, A, Quistgaard, E.M, Loew, C. | | Deposit date: | 2018-06-13 | | Release date: | 2019-01-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of Prototypic Peptide Transporter DtpA from E. coli in Complex with Valganciclovir Provides Insights into Drug Binding of Human PepT1.
J. Am. Chem. Soc., 141, 2019
|
|
8QW4
 
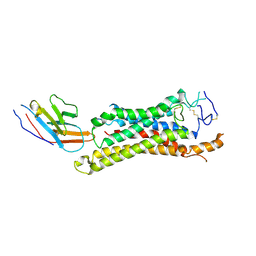 | | FZD3 in complex with nanobody 9 | | Descriptor: | Frizzled-3, Nanobody Nb9 | | Authors: | Zhao, Y, Jones, E.Y. | | Deposit date: | 2023-10-18 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into Frizzled3 through nanobody modulators.
Nat Commun, 15, 2024
|
|
4CGZ
 
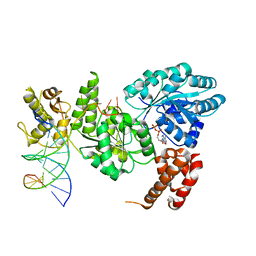 | | Crystal structure of the Bloom's syndrome helicase BLM in complex with DNA | | Descriptor: | 5'-D(*AP*GP*CP*GP*TP*CP*GP*AP*GP*AP*TP*CP)-3', 5'-D(*GP*AP*TP*CP*TP*CP*GP*AP*CP*GP*CP*TP*CP*DT *CP*CP*CP)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Newman, J.A, Savitsky, P, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2013-11-27 | | Release date: | 2013-12-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal Structure of the Bloom'S Syndrome Helicase Indicates a Role for the Hrdc Domain in Conformational Changes.
Nucleic Acids Res., 43, 2015
|
|
6QWL
 
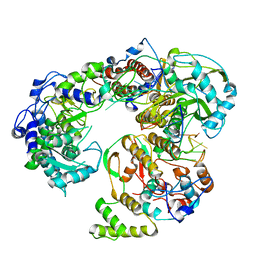 | | Influenza B virus (B/Panama/45) polymerase Hetermotrimer in complex with 3'5' cRNA promoter | | Descriptor: | 3' cRNA, 5' cRNA, Polymerase acidic protein, ... | | Authors: | Keown, J.R, Carrique, L, Fan, H, Fodor, E, Grimes, J.M. | | Deposit date: | 2019-03-05 | | Release date: | 2019-09-04 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structures of influenza A virus RNA polymerase offer insight into viral genome replication.
Nature, 573, 2019
|
|
6QPF
 
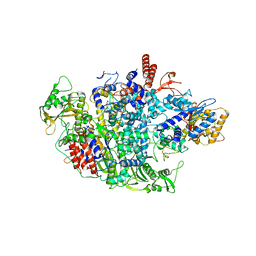 | | Influenza A virus Polymerase Heterotrimer A/duck/Fujian/01/2002(H5N1) | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2, RNA-directed RNA polymerase catalytic subunit | | Authors: | Fan, H.T, Keown, J.R, Fodor, E, Grimes, J.M. | | Deposit date: | 2019-02-13 | | Release date: | 2019-09-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.634 Å) | | Cite: | Structures of influenza A virus RNA polymerase offer insight into viral genome replication.
Nature, 573, 2019
|
|
5OCL
 
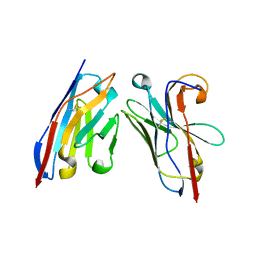 | | Nanobody-anti-VGLUT nanobody complex | | Descriptor: | anti-llama nanobody, anti-vglut nanobody | | Authors: | Dutzler, R, Schenck, S, Kunz, L. | | Deposit date: | 2017-07-03 | | Release date: | 2017-07-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Generation and Characterization of Anti-VGLUT Nanobodies Acting as Inhibitors of Transport.
Biochemistry, 56, 2017
|
|
6QX3
 
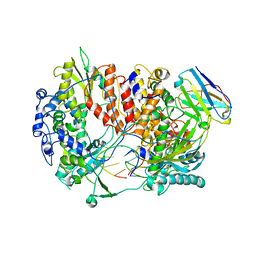 | | Influenza A virus (A/NT/60/1968) polymerase Hetermotrimer in complex with 3'5' cRNA promoter and Nb8205 | | Descriptor: | Nb8205, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Carrique, L, Keown, J.R, Fan, H, Fodor, E, Grimes, J.M. | | Deposit date: | 2019-03-07 | | Release date: | 2019-09-04 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.79 Å) | | Cite: | Structures of influenza A virus RNA polymerase offer insight into viral genome replication.
Nature, 573, 2019
|
|
6RR7
 
 | | Influenza A virus (A/NT/60/1968) polymerase Heterotrimer bound to 3'5' vRNA promoter and capped RNA primer | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2, RNA (5'-D(*(M7G))-R(P*AP*AP*UP*CP*U)-3'), ... | | Authors: | Carrique, L, Keown, J.R, Fan, H, Fodor, E, Grimes, J.M. | | Deposit date: | 2019-05-17 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structures of influenza A virus RNA polymerase offer insight into viral genome replication.
Nature, 573, 2019
|
|
6QPG
 
 | | Influenza A virus Polymerase Heterotrimer A/nt/60/1968(H3N2) in complex with Nanobody NB8205 | | Descriptor: | Nanobody NB8205, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Fan, H.T, Keown, J.R, Fodor, E, Grimes, J.M. | | Deposit date: | 2019-02-13 | | Release date: | 2019-09-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.34 Å) | | Cite: | Structures of influenza A virus RNA polymerase offer insight into viral genome replication.
Nature, 573, 2019
|
|
6QX8
 
 | | Influenza A virus (A/NT/60/1968) polymerase dimer of heterotrimer in complex with 5' cRNA promoter | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2, RNA (5'-R(P*AP*GP*CP*AP*AP*AP*AP*GP*CP*AP*GP*A)-3'), ... | | Authors: | Carrique, L, Keown, J.R, Fan, H, Fodor, E, Grimes, J.M. | | Deposit date: | 2019-03-07 | | Release date: | 2019-09-04 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.07 Å) | | Cite: | Structures of influenza A virus RNA polymerase offer insight into viral genome replication.
Nature, 573, 2019
|
|
6QXE
 
 | | Influenza A virus (A/NT/60/1968) polymerase dimer of hetermotrimer in complex with 3'5' cRNA promoter and Nb8205 | | Descriptor: | Nb8205, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Carrique, L, Keown, J.R, Fan, H, Fodor, E, Grimes, J.M. | | Deposit date: | 2019-03-07 | | Release date: | 2019-09-04 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.15 Å) | | Cite: | Structures of influenza A virus RNA polymerase offer insight into viral genome replication.
Nature, 573, 2019
|
|
8BE4
 
 | |
