4E4U
 
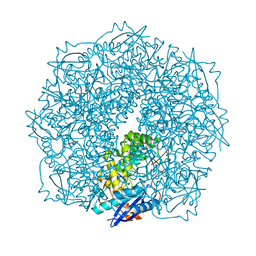 | | Crystal structure of a putative Mandelate racemase/Muconate lactonizing enzyme (Target PSI-200780) from Burkholderia SAR-1 | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, Mandalate racemase/muconate lactonizing enzyme | | Authors: | Kumar, P.R, Bonanno, J, Chowdhury, S, Foti, R, Gizzi, A, Hammonds, J, Hillerich, B, Matikainen, B, Seidel, R, Toro, R, Zencheck, W, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-03-13 | | Release date: | 2012-04-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of a putative MR/ML enzyme from Burkholderia SAR-1
to be published
|
|
4E4S
 
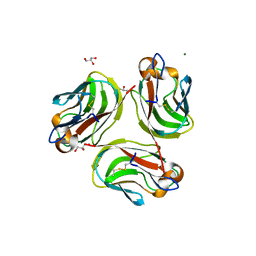 | | Crystal structure of Pika GITRL | | Descriptor: | GLYCEROL, MAGNESIUM ION, Tumor necrosis factor ligand superfamily member 18 | | Authors: | Kumar, P.R, Bhosle, R, Bonanno, J, Chowdhury, S, Gizzi, A, Glen, S, Hillerich, B, Hammonds, J, Seidel, R, Toro, R, Nathenson, S.G, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2012-03-13 | | Release date: | 2012-03-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of GITRL from Ochotona princeps
to be published
|
|
4E5T
 
 | | Crystal structure of a putative Mandelate racemase/Muconate lactonizing enzyme (Target PSI-200750) from Labrenzia alexandrii DFL-11 | | Descriptor: | MAGNESIUM ION, Mandelate racemase / muconate lactonizing enzyme, C-terminal domain protein | | Authors: | Kumar, P.R, Bonanno, J, Chowdhury, S, Foti, R, Gizzi, A, Glen, S, Hammonds, J, Hillerich, B, Matikainen, B, Seidel, R, Toro, R, Zencheck, W, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-03-14 | | Release date: | 2012-03-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of a putative MR/ML enzyme from Labrenzia alexandrii DFL-11
to be published
|
|
5TX7
 
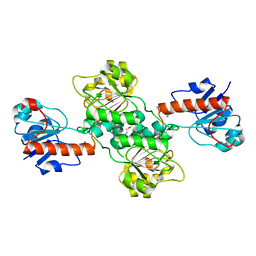 | | Crystal structure of D-isomer specific 2-hydroxyacid dehydrogenase from Desulfovibrio vulgaris | | Descriptor: | D-isomer specific 2-hydroxyacid dehydrogenase family protein, DI(HYDROXYETHYL)ETHER, TRIETHYLENE GLYCOL | | Authors: | Czub, M.P, Shabalin, I.G, Gasiorowska, O.A, Handing, K.B, Kutner, J, Cymborowski, M.T, Hennig, P.M, Bonanno, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2016-11-15 | | Release date: | 2016-12-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystal structure of D-isomer specific 2-hydroxyacid dehydrogenase from Desulfovibrio vulgaris
to be published
|
|
4AYR
 
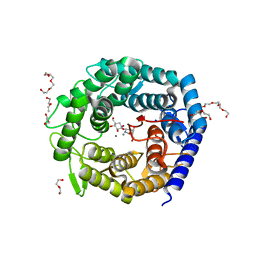 | | Structure of The GH47 processing alpha-1,2-mannosidase from Caulobacter strain K31 in complex with noeuromycin | | Descriptor: | (3S,4R,5R)-3,4-DIHYDROXY-5-(HYDROXYMETHYL)PIPERIDIN-2-ONE, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, CALCIUM ION, ... | | Authors: | Thompson, A.J, Dabin, J, Iglesias-Fernandez, J, Iglesias-Fernandez, A, Dinev, Z, Williams, S.J, Siriwardena, A, Moreland, C, Hu, T.C, Smith, D.K, Gilbert, H.J, Rovira, C, Davies, G.J. | | Deposit date: | 2012-06-21 | | Release date: | 2013-01-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The Reaction Coordinate of a Bacterial Gh47 Alpha-Mannosidase: A Combined Quantum Mechanical and Structural Approach.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
4AYP
 
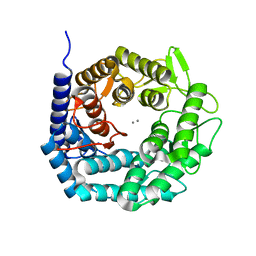 | | Structure of The GH47 processing alpha-1,2-mannosidase from Caulobacter strain K31 in complex with thiomannobioside | | Descriptor: | CALCIUM ION, MANNOSYL-OLIGOSACCHARIDE 1,2-ALPHA-MANNOSIDASE, SODIUM ION, ... | | Authors: | Thompson, A.J, Dabin, J, Iglesias-Fernandez, J, Iglesias-Fernandez, A, Dinev, Z, Williams, S.J, Siriwardena, A, Moreland, C, Hu, T.C, Smith, D.K, Gilbert, H.J, Rovira, C, Davies, G.J. | | Deposit date: | 2012-06-21 | | Release date: | 2013-01-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | The Reaction Coordinate of a Bacterial Gh47 Alpha-Mannosidase: A Combined Quantum Mechanical and Structural Approach.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
4AYO
 
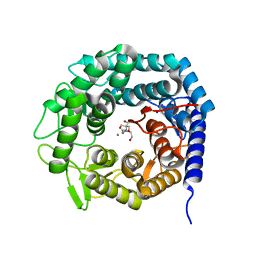 | | Structure of The GH47 processing alpha-1,2-mannosidase from Caulobacter strain K31 | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, MANNOSYL-OLIGOSACCHARIDE 1,2-ALPHA-MANNOSIDASE, ... | | Authors: | Thompson, A.J, Dabin, J, Iglesias-Fernandez, J, Iglesias-Fernandez, A, Dinev, Z, Williams, S.J, Siriwardena, A, Moreland, C, Hu, T.C, Smith, D.K, Gilbert, H.J, Rovira, C, Davies, G.J. | | Deposit date: | 2012-06-21 | | Release date: | 2013-01-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | The Reaction Coordinate of a Bacterial Gh47 Alpha-Mannosidase: A Combined Quantum Mechanical and Structural Approach.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
4AYQ
 
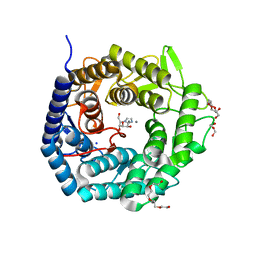 | | Structure of The GH47 processing alpha-1,2-mannosidase from Caulobacter strain K31 in complex with mannoimidazole | | Descriptor: | (5R,6R,7S,8R)-5-(HYDROXYMETHYL)-5,6,7,8-TETRAHYDROIMIDAZO[1,2-A]PYRIDINE-6,7,8-TRIOL, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Thompson, A.J, Dabin, J, Iglesias-Fernandez, J, Iglesias-Fernandez, A, Dinev, Z, Williams, S.J, Siriwardena, A, Moreland, C, Hu, T.C, Smith, D.K, Gilbert, H.J, Rovira, C, Davies, G.J. | | Deposit date: | 2012-06-21 | | Release date: | 2013-01-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The Reaction Coordinate of a Bacterial Gh47 Alpha-Mannosidase: A Combined Quantum Mechanical and Structural Approach.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
4XA8
 
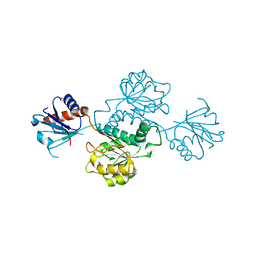 | | Crystal structure of D-isomer specific 2-hydroxyacid dehydrogenase from Xanthobacter autotrophicus Py2 | | Descriptor: | D-isomer specific 2-hydroxyacid dehydrogenase NAD-binding | | Authors: | Handing, K.B, Gasiorowska, O.A, Shabalin, I.G, Cymborowski, M.T, Bonanno, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-12-12 | | Release date: | 2015-01-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of D-isomer specific 2-hydroxyacid dehydrogenase from Xanthobacter autotrophicus Py2.
to be published
|
|
4XAP
 
 | | Crystal structure of aldo-keto reductase from Sinorhizobium meliloti 1021 | | Descriptor: | Aldo-keto reductase | | Authors: | Gasiorowska, O.A, Handing, K.B, Shabalin, I.G, Bonanno, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-12-15 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal structure of aldo-keto reductase from Sinorhizobium meliloti 1021
to be published
|
|
7Q4Q
 
 | | Magacizumab Fab fragment in complex with human LRG1 epitope | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, LRG1 epitope, Magacizumab heavy chain, ... | | Authors: | Gutierrez-Fernandez, J, Luecke, H. | | Deposit date: | 2021-11-01 | | Release date: | 2022-06-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis of human LRG1 recognition by Magacizumab, a humanized monoclonal antibody with therapeutic potential.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
4MDC
 
 | | Crystal structure of glutathione S-transferase from Sinorhizobium meliloti 1021, NYSGRC target 021389 | | Descriptor: | GLYCEROL, Putative glutathione S-transferase | | Authors: | Shabalin, I.G, Bacal, P, Cooper, D.R, Stead, M, Ahmed, M, Hammonds, J, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-08-22 | | Release date: | 2013-09-04 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of glutathione S-transferase from Sinorhizobium meliloti 1021
To be Published
|
|
4O9K
 
 | | Crystal structure of the CBS pair of a putative D-arabinose 5-phosphate isomerase from Methylococcus capsulatus in complex with CMP-Kdo | | Descriptor: | Arabinose 5-phosphate isomerase, CYTIDINE 5'-MONOPHOSPHATE 3-DEOXY-BETA-D-GULO-OCT-2-ULO-PYRANOSONIC ACID, GLYCEROL | | Authors: | Shabalin, I.G, Cooper, D.R, Shumilin, I.A, Zimmerman, M.D, Majorek, K.A, Hammonds, J, Hillerich, B.S, Nawar, A, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-01-02 | | Release date: | 2014-01-22 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure and kinetic properties of D-arabinose 5-phosphate isomerase from Methylococcus capsulatus
To be Published
|
|
4N18
 
 | | Crystal structure of D-isomer specific 2-hydroxyacid dehydrogenase family protein from Klebsiella pneumoniae 342 | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, CITRIC ACID, D-isomer specific 2-hydroxyacid dehydrogenase family protein | | Authors: | Bacal, P, Shabalin, I.G, Cooper, D.R, Majorek, K.A, Osinski, T, Hillerich, B.S, Hammonds, J, Nawar, A, Stead, M, Chowdhury, S, Gizzi, A, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-10-03 | | Release date: | 2013-10-16 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal structure of D-isomer specific 2-hydroxyacid dehydrogenase family protein from Klebsiella pneumoniae 342
To be Published
|
|
5BP9
 
 | | Crystal structure of SAM-dependent methyltransferase from Bacteroides fragilis in complex with S-Adenosyl-L-homocysteine | | Descriptor: | 2-{2-[2-2-(METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, Putative methyltransferase protein, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Gasiorowska, O.A, Shabalin, I.G, Handing, K.B, Cymborowski, M.T, Mason, D.V, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2015-05-27 | | Release date: | 2015-06-10 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of SAM-dependent methyltransferase fromBacteroides fragilis in complex with S-Adenosyl-L-homocysteine
to be published
|
|
5C5I
 
 | | Crystal structure of NADP-dependent dehydrogenase from Rhodobacter sphaeroides | | Descriptor: | NADP-dependent dehydrogenase | | Authors: | Kowiel, M, Gasiorowska, O.A, Shabalin, I.G, Handing, K.B, Porebski, P.J, Cymborowski, M, Al Obaidi, N.F, Bonanno, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2015-06-19 | | Release date: | 2015-07-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of NADP-dependent dehydrogenase from Rhodobacter sphaeroides
to be published
|
|
5C7H
 
 | | Crystal structure of aldo-keto reductase from Sinorhizobium meliloti 1021 in complex with NADPH | | Descriptor: | Aldo-keto reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Gasiorowska, O.A, Shabalin, I.G, Handing, K.B, Seidel, R, Bonanno, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2015-06-24 | | Release date: | 2015-07-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of aldo-keto reductase from Sinorhizobium meliloti 1021 in complex with NADPH
to be published
|
|
5BXY
 
 | | Crystal structure of RNA methyltransferase from Salinibacter ruber in complex with S-Adenosyl-L-homocysteine | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, RNA methyltransferase, ... | | Authors: | Handing, K.B, LaRowe, C, Shabalin, I.G, Stead, M, Hillerich, B.S, Ahmed, M, Bonanno, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2015-06-09 | | Release date: | 2015-07-01 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of RNA methylase family protein from Salinibacterruber in complex with S-Adenosyl-L-homocysteine.
to be published
|
|
4BE4
 
 | | Closed conformation of O. piceae sterol esterase | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gutierrez-Fernandez, J, Vaquero, M.E, Prieto, A, Barriuso, J, Gonzalez, M.J, Hermoso, J.A. | | Deposit date: | 2013-03-05 | | Release date: | 2014-03-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structures of Ophiostoma Piceae Sterol Esterase: Structural Insights Into Activation Mechanism and Product Release.
J.Struct.Biol., 187, 2014
|
|
3M7V
 
 | | Crystal structure of phosphopentomutase from streptococcus mutans | | Descriptor: | GLYCEROL, MANGANESE (II) ION, Phosphopentomutase | | Authors: | Fedorov, A.A, Bonanno, J, Fedorov, E.V, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-03-17 | | Release date: | 2010-04-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of phosphopentomutase from streptococcus mutans
To be Published
|
|
4BE9
 
 | | Open conformation of O. piceae sterol esterase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Gutierrez-Fernandez, J, Vaquero, M.E, Prieto, A, Barriuso, J, Gonzalez, M.J, Hermoso, J.A. | | Deposit date: | 2013-03-06 | | Release date: | 2014-03-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Ophiostoma Piceae Sterol Esterase: Structural Insights Into Activation Mechanism and Product Release.
J.Struct.Biol., 187, 2014
|
|
4CD8
 
 | | The structure of GH113 beta-mannanase AaManA from Alicyclobacillus acidocaldarius in complex with ManMIm | | Descriptor: | (5R,6R,7S,8R)-5-(HYDROXYMETHYL)-5,6,7,8-TETRAHYDROIMIDAZO[1,2-A]PYRIDINE-6,7,8-TRIOL, ENDO-BETA-1,4-MANNANASE, beta-D-mannopyranose | | Authors: | Williams, R.J, Iglesias-Fernandez, J, Stepper, J, Jackson, A, Thompson, A.J, Lowe, E.C, White, J.M, Gilbert, H.J, Rovira, C, Davies, G.J, Williams, S.J. | | Deposit date: | 2013-10-30 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Combined Inhibitor Free-Energy Landscape and Structural Analysis Reports on the Mannosidase Conformational Coordinate.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4CD5
 
 | | The structure of GH26 beta-mannanase CjMan26C from Cellvibrio japonicus in complex with ManMIm | | Descriptor: | (5R,6R,7S,8R)-5-(HYDROXYMETHYL)-5,6,7,8-TETRAHYDROIMIDAZO[1,2-A]PYRIDINE-6,7,8-TRIOL, ENDO-1,4-BETA MANNANASE, PUTATIVE, ... | | Authors: | Williams, R.J, Iglesias-Fernandez, J, Stepper, J, Jackson, A, Thompson, A.J, Lowe, E.C, White, J.M, Gilbert, H.J, Rovira, C, Davies, G.J, Williams, S.J. | | Deposit date: | 2013-10-30 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Combined Inhibitor Free-Energy Landscape and Structural Analysis Reports on the Mannosidase Conformational Coordinate.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4CD7
 
 | | The structure of GH113 beta-mannanase AaManA from Alicyclobacillus acidocaldarius in complex with ManIFG and beta-1,4-mannobiose | | Descriptor: | 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, ENDO-BETA-1,4-MANNANASE, beta-D-mannopyranose, ... | | Authors: | Williams, R.J, Iglesias-Fernandez, J, Stepper, J, Jackson, A, Thompson, A.J, Lowe, E.C, White, J.M, Gilbert, H.J, Rovira, C, Davies, G.J, Williams, S.J. | | Deposit date: | 2013-10-30 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Combined Inhibitor Free-Energy Landscape and Structural Analysis Reports on the Mannosidase Conformational Coordinate.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4CD4
 
 | | The structure of GH26 beta-mannanase CjMan26C from Cellvibrio japonicus in complex with ManIFG | | Descriptor: | 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, ENDO-1,4-BETA MANNANASE, PUTATIVE, ... | | Authors: | Williams, R.J, Iglesias-Fernandez, J, Stepper, J, Jackson, A, Thompson, A.J, Lowe, E.C, White, J.M, Gilbert, H.J, Rovira, C, Davies, G.J, Williams, S.J. | | Deposit date: | 2013-10-30 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Combined Inhibitor Free-Energy Landscape and Structural Analysis Reports on the Mannosidase Conformational Coordinate.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
