4CD0
 
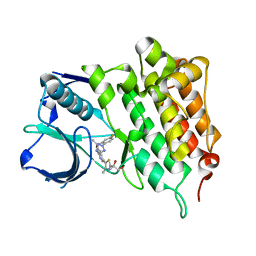 | | Structure of L1196M Mutant Human Anaplastic Lymphoma Kinase in Complex with 2-(5-(6-amino-5-((R)-1-(5-fluoro-2-(2H-1,2,3-triazol-2- yl)phenyl)ethoxy)pyridin-3-yl)-4-methylthiazol-2-yl)propane-1,2-diol | | Descriptor: | (2R)-2-[5-(6-amino-5-{(1R)-1-[2-(1,3-dihydro-2H-1,2,3-triazol-2-yl)-5-fluorophenyl]ethoxy}pyridin-3-yl)-4-methyl-1,3-thiazol-2-yl]propane-1,2-diol, ALK TYROSINE KINASE RECEPTOR | | Authors: | McTigue, M, Deng, Y, Liu, W, Brooun, A, Stewart, A. | | Deposit date: | 2013-10-29 | | Release date: | 2014-01-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | The Design of Potent and Selective Inhibitors to Overcome Clinical Alk Mutations Resistant to Crizotinib.
J.Med.Chem., 57, 2014
|
|
9BEW
 
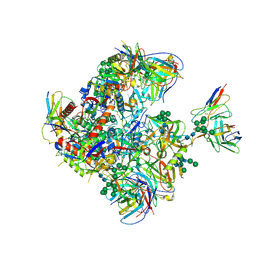 | |
9BER
 
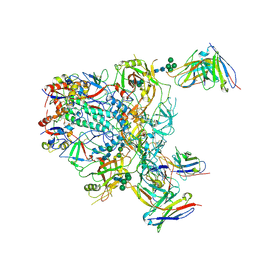 | | Cryo-EM structure of the HIV-1 JR-FL IDL Env trimer in complex with PGT122 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Gorman, J, Kwong, P.D. | | Deposit date: | 2024-04-16 | | Release date: | 2024-07-31 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Design of soluble HIV-1 envelope trimers free of covalent gp120-gp41 bonds with prevalent native-like conformation.
Cell Rep, 43, 2024
|
|
9BF6
 
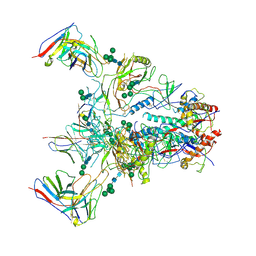 | |
3CK4
 
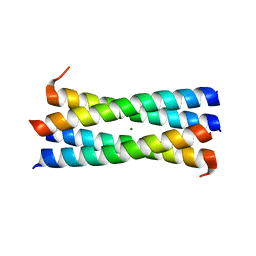 | | A heterospecific leucine zipper tetramer | | Descriptor: | GCN4 leucine zipper, MAGNESIUM ION | | Authors: | Liu, J. | | Deposit date: | 2008-03-14 | | Release date: | 2008-10-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A heterospecific leucine zipper tetramer.
Chem.Biol., 15, 2008
|
|
2RF2
 
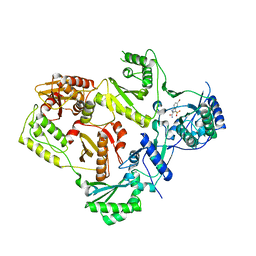 | | HIV reverse transcriptase in complex with inhibitor 7e (NNRTI) | | Descriptor: | 5-bromo-3-(pyrrolidin-1-ylsulfonyl)-1H-indole-2-carboxamide, Reverse transcriptase/ribonuclease H (EC 2.7.7.49) (EC 2.7.7.7) (EC 3.1.26.4) (p66 RT) | | Authors: | Yan, Y, Prasad, S. | | Deposit date: | 2007-09-27 | | Release date: | 2008-01-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Novel indole-3-sulfonamides as potent HIV non-nucleoside reverse transcriptase inhibitors (NNRTIs).
Bioorg.Med.Chem.Lett., 18, 2008
|
|
6RZ3
 
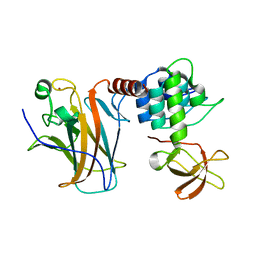 | | Crystal structure of a complex between the DNA-binding domain of p53 and the carboxyl-terminal conserved region of iASPP | | Descriptor: | Cellular tumor antigen p53, RelA-associated inhibitor, ZINC ION | | Authors: | Chen, S, Ren, J, Jones, E.Y, Lu, X. | | Deposit date: | 2019-06-12 | | Release date: | 2019-10-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (4.23 Å) | | Cite: | iASPP mediates p53 selectivity through a modular mechanism fine-tuning DNA recognition.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
7XK8
 
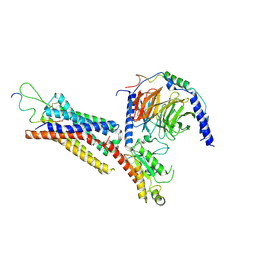 | | Cryo-EM structure of the Neuromedin U receptor 2 (NMUR2) in complex with G Protein and its endogeneous Peptide-Agonist NMU25 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Zhao, W, Wenru, Z, Mu, W, Minmin, L, Shutian, C, Tingting, T, Gisela, S, Holger, W, Albert, B, Cuiying, Y, Xiaojing, C, Han, S, Wu, B, Zhao, Q. | | Deposit date: | 2022-04-19 | | Release date: | 2023-02-22 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Ligand recognition and activation of neuromedin U receptor 2.
Nat Commun, 13, 2022
|
|
9KNY
 
 | | Cryo-EM structure of pyruvate-treated human mitochondrial pyruvate carrier in the IMS-open conformation at pH 8.0 | | Descriptor: | 1,2-DIOCTANOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CARDIOLIPIN, MPC specific nanobody 1, ... | | Authors: | Shi, J.H, Liang, J.M, Ma, D. | | Deposit date: | 2024-11-19 | | Release date: | 2025-03-12 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures and mechanism of the human mitochondrial pyruvate carrier.
Nature, 641, 2025
|
|
9KNW
 
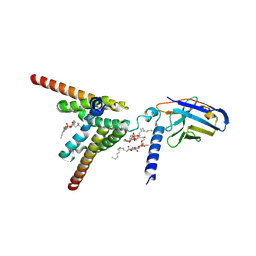 | | Cryo-EM structure of apo human mitochondrial pyruvate carrier in the IMS-open conformation at pH 6.8 | | Descriptor: | 1,2-DIOCTANOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CARDIOLIPIN, MPC specific nanobody 1, ... | | Authors: | Shi, J.H, Liang, J.M, Ma, D. | | Deposit date: | 2024-11-19 | | Release date: | 2025-03-12 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Structures and mechanism of the human mitochondrial pyruvate carrier.
Nature, 641, 2025
|
|
9KNX
 
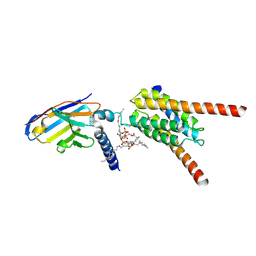 | | Cryo-EM structure of human mitochondrial pyruvate carrier in the occluded conformation at pH 6.8 | | Descriptor: | CARDIOLIPIN, MPC specific nanobody 1, Mitochondrial pyruvate carrier 1, ... | | Authors: | Shi, J.H, Liang, J.M, Ma, D. | | Deposit date: | 2024-11-19 | | Release date: | 2025-03-12 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Structures and mechanism of the human mitochondrial pyruvate carrier.
Nature, 641, 2025
|
|
7CJF
 
 | | Crystal structure of SARS-CoV-2 RBD in complex with a neutralizing antibody Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody heavy chain, ... | | Authors: | Guo, Y, Li, X, Zhang, G, Fu, D, Schweizer, L, Zhang, H, Rao, Z. | | Deposit date: | 2020-07-10 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.108 Å) | | Cite: | A SARS-CoV-2 neutralizing antibody with extensive Spike binding coverage and modified for optimal therapeutic outcomes.
Nat Commun, 12, 2021
|
|
8YW6
 
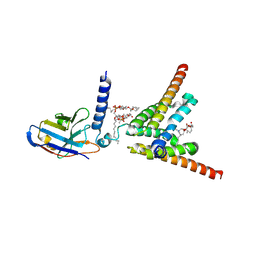 | | Cryo-EM structure of apo human mitochondrial pyruvate carrier in the IMS-open conformation at pH 8.0 | | Descriptor: | 1,2-DIOCTANOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CARDIOLIPIN, MPC specific nanobody 1, ... | | Authors: | Shi, J.H, Liang, J.M, Ma, D. | | Deposit date: | 2024-03-29 | | Release date: | 2025-03-12 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Structures and mechanism of the human mitochondrial pyruvate carrier.
Nature, 641, 2025
|
|
8YW9
 
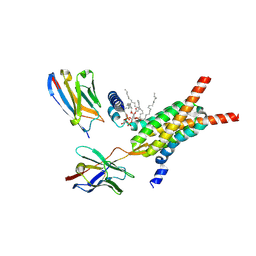 | | Cryo-EM structure of human mitochondrial pyruvate carrier in the matrix-facing conformation at pH 6.8 | | Descriptor: | CARDIOLIPIN, MPC specific nanobody 1, MPC specific nanobody 2, ... | | Authors: | Shi, J.H, Liang, J.M, Ma, D. | | Deposit date: | 2024-03-30 | | Release date: | 2025-03-12 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structures and mechanism of the human mitochondrial pyruvate carrier.
Nature, 641, 2025
|
|
8YW8
 
 | | Cryo-EM structure of human mitochondrial pyruvate carrier in complex with the inhibitor UK5099 | | Descriptor: | (E)-2-cyano-3-(1-phenylindol-3-yl)prop-2-enoic acid, CARDIOLIPIN, MPC specific nanobody 1, ... | | Authors: | Shi, J.H, Liang, J.M, Ma, D. | | Deposit date: | 2024-03-30 | | Release date: | 2025-03-12 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Structures and mechanism of the human mitochondrial pyruvate carrier.
Nature, 641, 2025
|
|
6URC
 
 | | Crystal structure of IRE1a in complex with compound 18 | | Descriptor: | 2-chloro-N-(6-methyl-5-{[3-(2-{[(3S)-piperidin-3-yl]amino}pyrimidin-4-yl)pyridin-2-yl]oxy}naphthalen-1-yl)benzene-1-sulfonamide, GLYCEROL, Serine/threonine-protein kinase/endoribonuclease IRE1 | | Authors: | Wallweber, H.H, Wang, W. | | Deposit date: | 2019-10-23 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Disruption of IRE1 alpha through its kinase domain attenuates multiple myeloma.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
9CZ7
 
 | | Crystal structure of integrin avb6 headpiece in complex with compound 12 | | Descriptor: | (2S)-phenyl{(3S)-3-[4-(5,6,7,8-tetrahydro-1,8-naphthyridin-2-yl)butoxy]pyrrolidin-1-yl}acetic acid, 17E6 Fab heavy chain, 17E6 Fab light chain, ... | | Authors: | Monroy, M.F, Qiao, Q, Lin, F.Y. | | Deposit date: | 2024-08-04 | | Release date: | 2024-11-13 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | The Discovery of MORF-627, a Highly Selective Conformationally-Biased Zwitterionic Integrin alpha v beta 6 Inhibitor for Fibrosis.
J.Med.Chem., 67, 2024
|
|
9CZF
 
 | | Crystal structure of integrin avb6 headpiece in complex with compound MORF-627 | | Descriptor: | (2S)-{5-fluoro-2-[(6S)-5-oxaspiro[2.5]octan-6-yl]phenyl}{(3R)-3-[4-(5,6,7,8-tetrahydro-1,8-naphthyridin-2-yl)butoxy]pyrrolidin-1-yl}acetic acid, 17E6 Fab heavy chain, 17E6 Fab light chain, ... | | Authors: | Monroy, M.F, Qiao, Q, Lin, F.Y. | | Deposit date: | 2024-08-05 | | Release date: | 2024-11-13 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | The Discovery of MORF-627, a Highly Selective Conformationally-Biased Zwitterionic Integrin alpha v beta 6 Inhibitor for Fibrosis.
J.Med.Chem., 67, 2024
|
|
9CZD
 
 | | Crystal structure of integrin avb6 headpiece in complex with compound 30 | | Descriptor: | (2S)-{5-fluoro-2-[(2S)-oxan-2-yl]phenyl}{(3R)-3-[4-(5,6,7,8-tetrahydro-1,8-naphthyridin-2-yl)butoxy]pyrrolidin-1-yl}acetic acid, 17E6 Fab heavy chain, 17E6 Fab light chain, ... | | Authors: | Monroy, M.F, Qiao, Q, Lin, F.Y. | | Deposit date: | 2024-08-05 | | Release date: | 2024-11-13 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | The Discovery of MORF-627, a Highly Selective Conformationally-Biased Zwitterionic Integrin alpha v beta 6 Inhibitor for Fibrosis.
J.Med.Chem., 67, 2024
|
|
9CZA
 
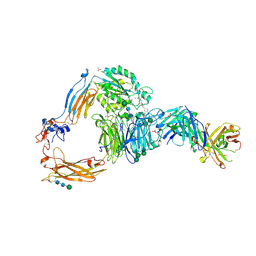 | | Crystal structure of integrin avb6 headpiece in complex with compound 18 | | Descriptor: | (2S)-(2-cyclopropylphenyl){(3R)-3-[4-(5,6,7,8-tetrahydro-1,8-naphthyridin-2-yl)butoxy]pyrrolidin-1-yl}acetic acid, 17E6 Fab heavy chain, 17E6 Fab light chain, ... | | Authors: | Monroy, M.F, Qiao, Q, Lin, F.Y. | | Deposit date: | 2024-08-05 | | Release date: | 2024-11-13 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | The Discovery of MORF-627, a Highly Selective Conformationally-Biased Zwitterionic Integrin alpha v beta 6 Inhibitor for Fibrosis.
J.Med.Chem., 67, 2024
|
|
3H42
 
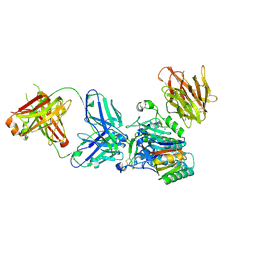 | | Crystal structure of PCSK9 in complex with Fab from LDLR competitive antibody | | Descriptor: | Fab from LDLR competitive antibody: Heavy chain, Fab from LDLR competitive antibody: Light chain, Proprotein convertase subtilisin/kexin type 9, ... | | Authors: | Piper, D.E, Walker, N.P.C, Romanow, W.G, Thibault, S.T, Tsai, M.M, Yang, E. | | Deposit date: | 2009-04-17 | | Release date: | 2009-05-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | From the Cover: A proprotein convertase subtilisin/kexin type 9 neutralizing antibody reduces serum cholesterol in mice and nonhuman primates.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
5BV7
 
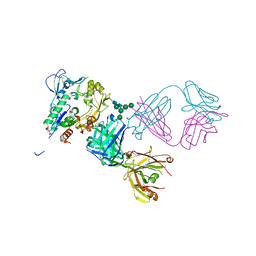 | | Crystal structure of human LCAT (L4F, N5D) in complex with Fab of an agonistic antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 27C3 heavy chain, 27C3 light chain, ... | | Authors: | Piper, D.E, Romanow, W.G, Thibault, S.T, Walker, N.P.C. | | Deposit date: | 2015-06-04 | | Release date: | 2015-12-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Agonistic Human Antibodies Binding to Lecithin-Cholesterol Acyltransferase Modulate High Density Lipoprotein Metabolism.
J.Biol.Chem., 291, 2016
|
|
8JAY
 
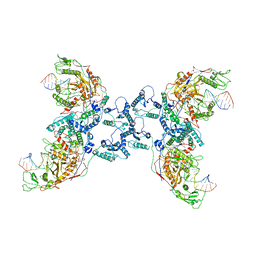 | | CrtSPARTA Octamer bound with guide-target | | Descriptor: | DNA (25-MER), MAGNESIUM ION, Piwi domain-containing protein, ... | | Authors: | Guo, L.J, Huang, P.P, Li, Z.X, Xiao, Y.B, Chen, M.R. | | Deposit date: | 2023-05-07 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Auto-inhibition and activation of a short Argonaute-associated TIR-APAZ defense system.
Nat.Chem.Biol., 20, 2024
|
|
7SLS
 
 | | HIV Reverse Transcriptase with compound Pyr02 | | Descriptor: | 5-(difluoromethyl)-3-{[1-{[(3S)-5-fluoro-2-methyl-6-oxo-3,6-dihydropyridin-3-yl]methyl}-6-oxo-4-(1,1,2,2-tetrafluoroethyl)-1,6-dihydropyrimidin-5-yl]oxy}-2-methylbenzonitrile, Reverse transcriptase/ribonuclease H | | Authors: | Klein, D.J, Zebisch, M, Gu, M. | | Deposit date: | 2021-10-24 | | Release date: | 2022-11-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.078 Å) | | Cite: | Potent targeted activator of cell kill molecules eliminate cells expressing HIV-1.
Sci Transl Med, 15, 2023
|
|
7SLR
 
 | | HIV Reverse Transcriptase with compound Pyr01 | | Descriptor: | 5-(difluoromethyl)-3-({1-[(5-fluoro-2-oxo-1,2-dihydropyridin-3-yl)methyl]-6-oxo-4-(1,1,2,2-tetrafluoroethyl)-1,6-dihydropyrimidin-5-yl}oxy)-2-methylbenzonitrile, Reverse transcriptase/ribonuclease H | | Authors: | Klein, D.J, Zebisch, M, Gu, M. | | Deposit date: | 2021-10-24 | | Release date: | 2022-11-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.179 Å) | | Cite: | Potent targeted activator of cell kill molecules eliminate cells expressing HIV-1.
Sci Transl Med, 15, 2023
|
|
