8ZM8
 
 | | Crystal Structure of the first bromodomain of human BRD4 BD1 in complex with the inhibitor Y13221 | | Descriptor: | 2-[2-ethyl-4-(methoxymethyl)-1~{H}-imidazol-5-yl]-7-[2-(4-fluoranyl-2,6-dimethyl-phenoxy)-5-(2-oxidanylpropan-2-yl)phenyl]-5-methyl-furo[3,2-c]pyridin-4-one, BRD4_HUMAN | | Authors: | Li, J, Hu, Q, Xu, H, Zhao, X, Zhang, C, Zhu, R, Wu, X, Zhang, Y, Xu, Y. | | Deposit date: | 2024-05-22 | | Release date: | 2024-12-11 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Discovery of the First BRD4 Second Bromodomain (BD2)-Selective Inhibitors.
J.Med.Chem., 67, 2024
|
|
8ZMB
 
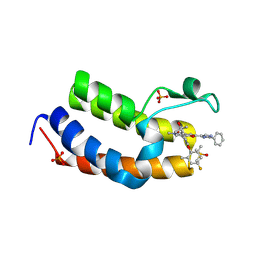 | | Crystal Structure of the first bromodomain of human BRD4 BD1 in complex with the inhibitor Y13195 | | Descriptor: | 7-(2-(4-fluoro-2,6-dimethylphenoxy)-5-(2-hydroxypropan-2-yl)phenyl)-5-methyl-2-(2-phenyl-1H-imidazol-5-yl)furo[3,2-c]pyridin-4(5H)-one, BRD4_HUMAN, DIMETHYL SULFOXIDE, ... | | Authors: | Li, J, Hu, Q, Xu, H, Zhao, X, Zhang, C, Zhu, R, Wu, X, Zhang, Y, Xu, Y. | | Deposit date: | 2024-05-22 | | Release date: | 2024-12-11 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Discovery of the First BRD4 Second Bromodomain (BD2)-Selective Inhibitors.
J.Med.Chem., 67, 2024
|
|
4JOC
 
 | |
4JOB
 
 | |
7C2Z
 
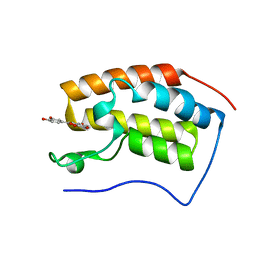 | | Bromodomain-containing 4 BD1 in complex with 3',4',7,8-Tetrahydroxyflavone | | Descriptor: | 2-[3,4-bis(oxidanyl)phenyl]-7,8-bis(oxidanyl)chromen-4-one, Bromodomain-containing protein 4, FORMIC ACID | | Authors: | Li, J, Zhu, J. | | Deposit date: | 2020-05-10 | | Release date: | 2021-05-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Discovery of the natural product 3',4',7,8-tetrahydroxyflavone as a novel and potent selective BRD4 bromodomain 2 inhibitor.
J Enzyme Inhib Med Chem, 36, 2021
|
|
7C6P
 
 | | Bromodomain-containing 4 BD2 in complex with 3',4',7,8- Tetrahydroxyflavonoid | | Descriptor: | 2-[3,4-bis(oxidanyl)phenyl]-7,8-bis(oxidanyl)chromen-4-one, Bromodomain-containing protein 4 | | Authors: | Li, J, Yu, K, Luo, Y, Zheng, W, Liang, W, Zhu, J. | | Deposit date: | 2020-05-22 | | Release date: | 2021-05-12 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Discovery of the natural product 3',4',7,8-tetrahydroxyflavone as a novel and potent selective BRD4 bromodomain 2 inhibitor.
J Enzyme Inhib Med Chem, 36, 2021
|
|
8Z8W
 
 | |
5YIS
 
 | | Crystal Structure of AnkB LIR/LC3B complex | | Descriptor: | Ankyrin-2, GLYCEROL, Microtubule-associated proteins 1A/1B light chain 3B, ... | | Authors: | Li, J, Zhu, R, Chen, K, Zheng, H, Yuan, C, Zhang, H, Wang, C, Zhang, M. | | Deposit date: | 2017-10-06 | | Release date: | 2018-05-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Potent and specific Atg8-targeting autophagy inhibitory peptides from giant ankyrins.
Nat. Chem. Biol., 14, 2018
|
|
5YIR
 
 | | Crystal Structure of AnkB LIR/GABARAP complex | | Descriptor: | Ankyrin-2, Gamma-aminobutyric acid receptor-associated protein, NICKEL (II) ION | | Authors: | Li, J, Zhu, R, Chen, K, Zheng, H, Yuan, C, Zhang, H, Wang, C, Zhang, M. | | Deposit date: | 2017-10-06 | | Release date: | 2018-05-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Potent and specific Atg8-targeting autophagy inhibitory peptides from giant ankyrins.
Nat. Chem. Biol., 14, 2018
|
|
5YIQ
 
 | | Crystal structure of AnkG LIR/LC3B complex | | Descriptor: | Ankyrin-3, Microtubule-associated proteins 1A/1B light chain 3B, ZINC ION | | Authors: | Li, J, Zhu, R, Chen, K, Zheng, H, Yuan, C, Zhang, H, Wang, C, Zhang, M. | | Deposit date: | 2017-10-06 | | Release date: | 2018-05-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Potent and specific Atg8-targeting autophagy inhibitory peptides from giant ankyrins.
Nat. Chem. Biol., 14, 2018
|
|
5YIP
 
 | | Crystal Structure of AnkG LIR/GABARAPL1 complex | | Descriptor: | Ankyrin-3, GLYCEROL, Gamma-aminobutyric acid receptor-associated protein-like 1 | | Authors: | Li, J, Zhu, R, Chen, K, Zheng, H, Yuan, C, Zhang, H, Wang, C, Zhang, M. | | Deposit date: | 2017-10-06 | | Release date: | 2018-05-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Potent and specific Atg8-targeting autophagy inhibitory peptides from giant ankyrins.
Nat. Chem. Biol., 14, 2018
|
|
7CUQ
 
 | | 2.55-Angstrom Cryo-EM structure of Cytochrome bo3 from Escherichia coli in Native Membrane | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, COPPER (II) ION, Cytochrome bo(3) ubiquinol oxidase subunit 1, ... | | Authors: | Li, J, Han, L, Gennis, R.B, Zhu, J.P, Zhang, K. | | Deposit date: | 2020-08-24 | | Release date: | 2021-08-25 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Cryo-EM structures of Escherichia coli cytochrome bo3 reveal bound phospholipids and ubiquinone-8 in a dynamic substrate binding site.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CUB
 
 | | 2.55-Angstrom Cryo-EM structure of Cytochrome bo3 from Escherichia coli in Native Membrane | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, COPPER (II) ION, Cytochrome bo(3) ubiquinol oxidase subunit 1, ... | | Authors: | Li, J, Han, L, Gennis, R.B, Zhu, J.P, Zhang, K. | | Deposit date: | 2020-08-22 | | Release date: | 2021-08-25 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | Cryo-EM structures of Escherichia coli cytochrome bo3 reveal bound phospholipids and ubiquinone-8 in a dynamic substrate binding site.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CUW
 
 | | Ubiquinol Binding Site of Cytochrome bo3 from Escherichia coli | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, COPPER (II) ION, Cytochrome bo(3) ubiquinol oxidase subunit 1, ... | | Authors: | Li, J, Han, L, Gennis, R.B, Zhu, J.P, Zhang, K. | | Deposit date: | 2020-08-25 | | Release date: | 2021-08-25 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | Cryo-EM structures of Escherichia coli cytochrome bo3 reveal bound phospholipids and ubiquinone-8 in a dynamic substrate binding site.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8YCM
 
 | | Monomeric Human STK19 | | Descriptor: | Isoform 4 of Inactive serine/threonine-protein kinase 19, SULFATE ION | | Authors: | Li, J, Ma, X, Dong, Z. | | Deposit date: | 2024-02-18 | | Release date: | 2024-03-06 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Mutations found in cancer patients compromise DNA binding of the winged helix protein STK19.
Sci Rep, 14, 2024
|
|
7U20
 
 | | Crystal structure of human METTL1 and WDR4 complex | | Descriptor: | SULFATE ION, tRNA (guanine-N(7)-)-methyltransferase, tRNA (guanine-N(7)-)-methyltransferase non-catalytic subunit WDR4 | | Authors: | Li, J, Nowak, R.P, Fischer, E.S, Gregory, R. | | Deposit date: | 2022-02-22 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis of regulated m 7 G tRNA modification by METTL1-WDR4.
Nature, 613, 2023
|
|
7WI7
 
 | | Crystal structure of human MCM8/9 complex | | Descriptor: | DNA helicase MCM8, DNA helicase MCM9, ZINC ION | | Authors: | Li, J, Liu, Y. | | Deposit date: | 2022-01-03 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (6.6 Å) | | Cite: | Crystal structure of human MCM8/9 complex
To Be Published
|
|
7XCL
 
 | |
7XCM
 
 | | Crystal structure of sulfite MttB structure at 3.2 A resolution | | Descriptor: | 3-METHYL-5-SULFO-PYRROLIDINE-2-CARBOXYLIC ACID, GLYCEROL, SODIUM ION, ... | | Authors: | Li, J, Chan, M.K. | | Deposit date: | 2022-03-24 | | Release date: | 2023-01-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Insights into pyrrolysine function from structures of a trimethylamine methyltransferase and its corrinoid protein complex.
Commun Biol, 6, 2023
|
|
7XCN
 
 | | Crystal structure of the MttB-MttC complex at 2.7 A resolution | | Descriptor: | 5-HYDROXYBENZIMIDAZOLYLCOBAMIDE, GLYCEROL, Trimethylamine methyltransferase, ... | | Authors: | Li, J, Chan, M.K. | | Deposit date: | 2022-03-24 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Insights into pyrrolysine function from structures of a trimethylamine methyltransferase and its corrinoid protein complex.
Commun Biol, 6, 2023
|
|
7BV3
 
 | | Crystal structure of a ugt transferase from Siraitia grosvenorii in complex with UDP | | Descriptor: | Glycosyltransferase, URIDINE-5'-DIPHOSPHATE | | Authors: | Li, J, Shan, N, Yang, J.G, Liu, W.D, Sun, Y.X. | | Deposit date: | 2020-04-09 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Near-perfect control of the regioselective glucosylation enabled by rational design of glycosyltransferases
Green Synth Catal, 2021
|
|
7DPD
 
 | | Human MCM9 N-terminal domain | | Descriptor: | DNA helicase MCM9, SODIUM ION, ZINC ION | | Authors: | Li, J, Liu, L, Liu, Y. | | Deposit date: | 2020-12-18 | | Release date: | 2021-05-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural study of the N-terminal domain of human MCM8/9 complex.
Structure, 29, 2021
|
|
7DP3
 
 | | Human MCM8 N-terminal domain | | Descriptor: | DNA helicase MCM8, ZINC ION | | Authors: | Li, J, Liu, L, Liu, Y. | | Deposit date: | 2020-12-17 | | Release date: | 2021-05-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural study of the N-terminal domain of human MCM8/9 complex.
Structure, 29, 2021
|
|
7E29
 
 | | Crystal Structure of Saccharomyces cerevisiae Ioc4 PWWP domain fused with MBP | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,ISWI one complex protein 4, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Li, J, Smolle, M, Liang, H, Liu, Y. | | Deposit date: | 2021-02-05 | | Release date: | 2022-02-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | H3K36 methylation and DNA-binding both promote Ioc4 recruitment and Isw1b remodeler function.
Nucleic Acids Res., 50, 2022
|
|
4L94
 
 | | Crystal structure of Human Hsp90 with S46 | | Descriptor: | (4-hydroxyphenyl)(4-methylpiperazin-1-yl)methanone, Heat shock protein HSP 90-alpha | | Authors: | Li, J, Ren, J, Yang, M, Xiong, B, He, J. | | Deposit date: | 2013-06-18 | | Release date: | 2014-06-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.649 Å) | | Cite: | Identification of a new series of potent diphenol HSP90 inhibitors by fragment merging and structure-based optimization
Bioorg.Med.Chem.Lett., 24, 2014
|
|
