7WP9
 
 | | SARS-CoV-2 Omicron Variant SPIKE trimer, all RBDs down | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yin, W, Xu, Y, Xu, P, Cao, X, Wu, C, Gu, C, He, X, Wang, X, Huang, S, Yuan, Q, Wu, K, Hu, W, Huang, Z, Liu, J, Wang, Z, Jia, F, Xia, K, Liu, P, Wang, X, Song, B, Zheng, J, Jiang, H, Cheng, X, Jiang, Y, Deng, S, Xu, E. | | Deposit date: | 2022-01-23 | | Release date: | 2022-03-09 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Structures of the Omicron spike trimer with ACE2 and an anti-Omicron antibody.
Science, 375, 2022
|
|
2JR8
 
 | | Solution structure of Manduca sexta moricin | | Descriptor: | Antimicrobial peptide moricin | | Authors: | Gong, Y, Dai, H, Rayaprolu, S, Huang, R, Prakash, O, Jiang, H. | | Deposit date: | 2007-06-21 | | Release date: | 2008-03-25 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution structure, antibacterial activity, and expression profile of Manduca sexta moricin.
J.Pept.Sci., 14, 2008
|
|
7FIH
 
 | | luteinizing hormone/choriogonadotropin receptor(S277I)-chorionic gonadotropin-Gs-Org43553 complex | | Descriptor: | 5-azanyl-N-tert-butyl-2-methylsulfanyl-4-[3-(2-morpholin-4-ylethanoylamino)phenyl]thieno[2,3-d]pyrimidine-6-carboxamide, Choriogonadotropin subunit beta 3, Engineered Guanine nucleotide-binding protein G(s) subunit alpha, ... | | Authors: | Duan, J, Xu, P, Cheng, X, Mao, C, Croll, T, He, X, Shi, J, Luan, X, Yin, W, You, E, Liu, Q, Zhang, S, Jiang, H, Zhang, Y, Jiang, Y, Xu, H.E. | | Deposit date: | 2021-07-31 | | Release date: | 2021-09-29 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures of full-length glycoprotein hormone receptor signalling complexes.
Nature, 598, 2021
|
|
7FII
 
 | | luteinizing hormone/choriogonadotropin receptor-chorionic gonadotropin-Gs complex | | Descriptor: | Choriogonadotropin subunit beta 3, Engineered Guanine nucleotide-binding protein G(s) subunit alpha, Glycoprotein hormones alpha chain, ... | | Authors: | Duan, J, Xu, P, Cheng, X, Mao, C, Croll, T, He, X, Shi, J, Luan, X, Yin, W, You, E, Liu, Q, Zhang, S, Jiang, H, Zhang, Y, Jiang, Y, Xu, H.E. | | Deposit date: | 2021-07-31 | | Release date: | 2021-09-29 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structures of full-length glycoprotein hormone receptor signalling complexes.
Nature, 598, 2021
|
|
7FIG
 
 | | luteinizing hormone/choriogonadotropin receptor(S277I)-chorionic gonadotropin-Gs complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Camelid antibody VHH fragment Nb35, Choriogonadotropin subunit beta 3, ... | | Authors: | Duan, J, Xu, P, Cheng, X, Mao, C, Croll, T, He, X, Shi, J, Luan, X, Yin, W, You, E, Liu, Q, Zhang, S, Jiang, H, Zhang, Y, Jiang, Y, Xu, H.E. | | Deposit date: | 2021-07-31 | | Release date: | 2021-09-29 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structures of full-length glycoprotein hormone receptor signalling complexes.
Nature, 598, 2021
|
|
7FIJ
 
 | | luteinizing hormone/choriogonadotropin receptor | | Descriptor: | Lutropin-choriogonadotropic hormone receptor | | Authors: | Duan, J, Xu, P, Cheng, X, Mao, C, Croll, T, He, X, Shi, J, Luan, X, Yin, W, You, E, Liu, Q, Zhang, S, Jiang, H, Zhang, Y, Jiang, Y, Xu, H.E. | | Deposit date: | 2021-07-31 | | Release date: | 2021-09-29 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structures of full-length glycoprotein hormone receptor signalling complexes.
Nature, 598, 2021
|
|
5H9G
 
 | |
5H9H
 
 | |
8Y40
 
 | | Structure of chimeric RyR-I4657M/G4819E complex with chlorantraniliprole | | Descriptor: | 5-bromanyl-N-[4-chloranyl-2-methyl-6-(methylcarbamoyl)phenyl]-2-(3-chloranylpyridin-2-yl)pyrazole-3-carboxamide, ADENOSINE-5'-TRIPHOSPHATE, CAFFEINE, ... | | Authors: | Lin, L, Wang, C, Wang, W, Jiang, H, Yuchi, Z. | | Deposit date: | 2024-01-29 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Cryo-EM structures of ryanodine receptors and diamide insecticides reveal the mechanisms of selectivity and resistance.
Nat Commun, 15, 2024
|
|
8XKH
 
 | | Structure of chimeric RyR Complex with tetraniliprole | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CAFFEINE, CALCIUM ION, ... | | Authors: | Lin, L, Wang, C, Wang, W, Jiang, H, Yuchi, Z. | | Deposit date: | 2023-12-23 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | Cryo-EM structures of ryanodine receptors and diamide insecticides reveal the mechanisms of selectivity and resistance.
Nat Commun, 15, 2024
|
|
8XLH
 
 | | Structure of chimeric RyR-I4657M/G4819E | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CAFFEINE, CALCIUM ION, ... | | Authors: | Lin, L, Wang, C, Wang, W, Jiang, H, Yuchi, Z. | | Deposit date: | 2023-12-26 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Cryo-EM structures of ryanodine receptors and diamide insecticides reveal the mechanisms of selectivity and resistance.
Nat Commun, 15, 2024
|
|
8XJI
 
 | | Structure of chimeric RyR complex with flubendiamide | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CAFFEINE, CALCIUM ION, ... | | Authors: | Lin, L, Wang, C, Wang, W, Jiang, H, Yuchi, Z. | | Deposit date: | 2023-12-21 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.91 Å) | | Cite: | Cryo-EM structures of ryanodine receptors and diamide insecticides reveal the mechanisms of selectivity and resistance.
Nat Commun, 15, 2024
|
|
8XLF
 
 | | Structure of chimeric RyR | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CAFFEINE, CALCIUM ION, ... | | Authors: | Lin, L, Wang, C, Wang, W, Jiang, H, Yuchi, Z. | | Deposit date: | 2023-12-25 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Cryo-EM structures of ryanodine receptors and diamide insecticides reveal the mechanisms of selectivity and resistance.
Nat Commun, 15, 2024
|
|
5ZIU
 
 | | Crystal structure of human Entervirus D68 RdRp | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, RdRp | | Authors: | Wang, M.L, Zhang, Y, Chen, Y.P, Lu, D.R, Jiang, H, Chen, Y.J, Li, L, Zhang, C.H, Shi, Q.L, Su, D. | | Deposit date: | 2018-03-17 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.147 Å) | | Cite: | Crystal structure of human Entervirus D68 RdRp in complex with NADPH
To Be Published
|
|
8JRE
 
 | | Cryo-EM structure of a designed AAV8-based vector | | Descriptor: | Capsid protein | | Authors: | Ke, X, Luo, S, Zheng, Q, Jiang, H, Liu, F, Sun, X. | | Deposit date: | 2023-06-16 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.08 Å) | | Cite: | An adeno-associated virus variant enabling efficient ocular-directed gene delivery across species.
Nat Commun, 15, 2024
|
|
8YP2
 
 | | the crystal structure of inactive Magnaporthe grisea oxidoreductase in complex with NADP and Glycerol | | Descriptor: | GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADP-dependent oxidoreductase domain-containing protein | | Authors: | Huang, X, Jiang, H, Tang, D, Lin, S. | | Deposit date: | 2024-03-15 | | Release date: | 2025-03-19 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Crystal Structure of an Aldo-keto Reductase MGG_00097 from Magnaporthe grisea.
Plant Pathol J, 41, 2025
|
|
8YP9
 
 | | the crystal structure of wildtype Magnaporthe grisea oxidoreductase in complex with NADP | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADP-dependent oxidoreductase domain-containing protein | | Authors: | Huang, X, Jiang, H, Tang, D, Lin, S. | | Deposit date: | 2024-03-15 | | Release date: | 2025-03-19 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of an Aldo-keto Reductase MGG_00097 from Magnaporthe grisea.
Plant Pathol J, 41, 2025
|
|
7F16
 
 | | Cryo-EM structure of parathyroid hormone receptor type 2 in complex with a tuberoinfundibular peptide of 39 residues and G protein | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, X, Cheng, X, Zhao, L.H, Wang, Y, Ye, C, Zou, X, Dai, A, Cong, Z, Chen, J, Zhou, Q, Xia, T, Jiang, H, Xu, H.E, Yang, D, Wang, M.W. | | Deposit date: | 2021-06-08 | | Release date: | 2021-08-18 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Molecular insights into differentiated ligand recognition of the human parathyroid hormone receptor 2.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7F2E
 
 | | SARS-CoV-2 nucleocapsid protein C-terminal domain (dodecamer) | | Descriptor: | Nucleoprotein, PHOSPHATE ION | | Authors: | Liu, C, Jiang, H. | | Deposit date: | 2021-06-10 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structures of the SARS-CoV-2 nucleocapsid protein C-terminal domain and development of nucleocapsid-targeting nanobodies.
Febs J., 289, 2022
|
|
8HVL
 
 | | Crystal structure of SARS-Cov-2 main protease M49I mutant in complex with PF07321332 | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Zou, X.F, Zhang, J, Li, J. | | Deposit date: | 2022-12-27 | | Release date: | 2023-12-27 | | Last modified: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Evaluation of the Inhibition Potency of Nirmatrelvir against Main Protease Mutants of SARS-CoV-2 Variants.
Biochemistry, 62, 2023
|
|
8HVO
 
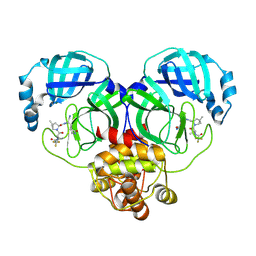 | | Crystal structure of SARS-Cov-2 main protease V186F mutant in complex with PF07321332 | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Zou, X.F, Zhang, J, Li, J. | | Deposit date: | 2022-12-27 | | Release date: | 2023-12-27 | | Last modified: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Evaluation of the Inhibition Potency of Nirmatrelvir against Main Protease Mutants of SARS-CoV-2 Variants.
Biochemistry, 62, 2023
|
|
8HVM
 
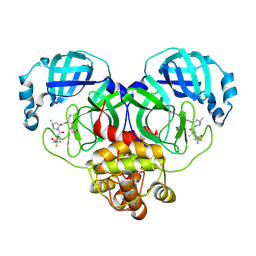 | | Crystal structure of SARS-Cov-2 main protease K90R mutant in complex with PF07321332 | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Wang, J, Zhang, J, Li, J. | | Deposit date: | 2022-12-27 | | Release date: | 2023-12-27 | | Last modified: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Evaluation of the Inhibition Potency of Nirmatrelvir against Main Protease Mutants of SARS-CoV-2 Variants.
Biochemistry, 62, 2023
|
|
8HVK
 
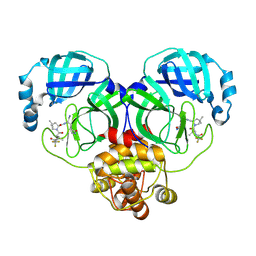 | | Crystal structure of SARS-Cov-2 main protease G15S mutant in complex with PF07321332 | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Zeng, P, Zhang, J, Li, J. | | Deposit date: | 2022-12-27 | | Release date: | 2023-12-27 | | Last modified: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Evaluation of the Inhibition Potency of Nirmatrelvir against Main Protease Mutants of SARS-CoV-2 Variants.
Biochemistry, 62, 2023
|
|
8HVN
 
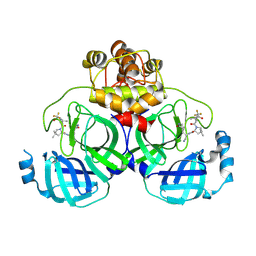 | | Crystal structure of SARS-Cov-2 main protease P132H mutant in complex with PF07321332 | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Li, W.W, Zhang, J, Li, J. | | Deposit date: | 2022-12-27 | | Release date: | 2023-12-27 | | Last modified: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Evaluation of the Inhibition Potency of Nirmatrelvir against Main Protease Mutants of SARS-CoV-2 Variants.
Biochemistry, 62, 2023
|
|
8HVU
 
 | | Crystal structure of SARS-Cov-2 main protease G15S mutant in complex with PF07304814 | | Descriptor: | 3C-like proteinase nsp5, [(3~{S})-3-[[(2~{S})-2-[(4-methoxy-1~{H}-indol-2-yl)carbonylamino]-4-methyl-pentanoyl]amino]-2-oxidanylidene-4-[(3~{R})-2-oxidanylidene-3,4-dihydropyrrol-3-yl]butyl] dihydrogen phosphate | | Authors: | Zeng, P, Zhang, J, Li, J. | | Deposit date: | 2022-12-28 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Crystal structures of main protease (M pro ) mutants of SARS-CoV-2 variants bound to PF-07304814.
Mol Biomed, 4, 2023
|
|
