3F4E
 
 | |
3BCC
 
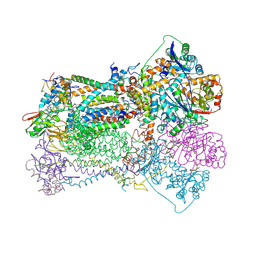 | | STIGMATELLIN AND ANTIMYCIN BOUND CYTOCHROME BC1 COMPLEX FROM CHICKEN | | Descriptor: | ANTIMYCIN, FE2/S2 (INORGANIC) CLUSTER, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Zhang, Z, Huang, L, Shulmeister, V.M, Chi, Y.-I, Kim, K.K, Hung, L.-W, Crofts, A.R, Berry, E.A, Kim, S.-H. | | Deposit date: | 1998-03-23 | | Release date: | 1998-08-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Electron transfer by domain movement in cytochrome bc1.
Nature, 392, 1998
|
|
4DKS
 
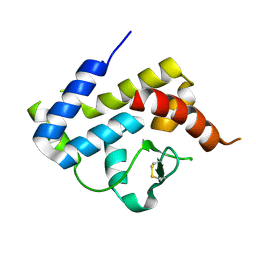 | | A spindle-shaped virus protein (chymotrypsin treated) | | Descriptor: | Probable integrase | | Authors: | Ouyang, S, Liang, W, Huang, L, Liu, Z.-J. | | Deposit date: | 2012-02-04 | | Release date: | 2012-05-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and functional characterization of the C-terminal catalytic domain of SSV1 integrase.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4E2P
 
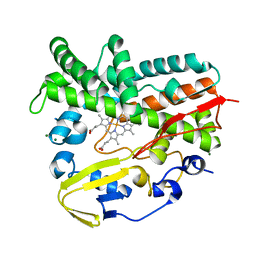 | | Crystal Structure of a Post-tailoring Hydroxylase (HmtN) Involved in the Himastatin Biosynthesis | | Descriptor: | Cytochrome P450 107B1 (P450CVIIB1), MAGNESIUM ION, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Zhang, H.D, Chen, J, Wang, H, Huang, L, Zhang, H.J. | | Deposit date: | 2012-03-09 | | Release date: | 2013-03-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Crystal Structure of a Post-tailoring Hydroxylase (HmtN) Involved in the Himastatin Biosynthesis
To be Published
|
|
2OJU
 
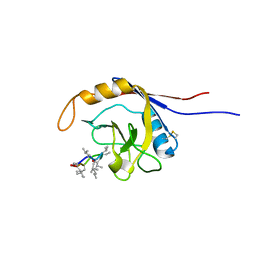 | | X-ray structure of complex of human cyclophilin J with cyclosporin A | | Descriptor: | CYCLOSPORIN A, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE-LIKE 3 | | Authors: | Xia, Z, Huang, L. | | Deposit date: | 2007-01-14 | | Release date: | 2008-01-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Targeting Cyclophilin J, a Novel Peptidyl-Prolyl Isomerase, Can Induce Cellular G1/S Arrest and Repress the Growth of Hepatocellular Carcinoma
To be Published
|
|
3VCF
 
 | | SSV1 integrase C-terminal catalytic domain (174-335aa) | | Descriptor: | Probable integrase | | Authors: | Ouyang, S, Liang, W, Huang, L, Liu, Z.-J. | | Deposit date: | 2012-01-04 | | Release date: | 2012-05-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and functional characterization of the C-terminal catalytic domain of SSV1 integrase.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
7CD9
 
 | | Crystal Structure of SETDB1 tudor domain in complexed with Compound 6 | | Descriptor: | 3-methyl-2-[[(3R,5R)-1-methyl-5-(4-phenylmethoxyphenyl)piperidin-3-yl]amino]-5H-pyrrolo[3,2-d]pyrimidin-4-one, CITRIC ACID, Histone-lysine N-methyltransferase SETDB1 | | Authors: | Xiong, L, Guo, Y, Mao, X, Huang, L, Wu, C, Yang, S. | | Deposit date: | 2020-06-19 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Guided Discovery of a Potent and Selective Cell-Active Inhibitor of SETDB1 Tudor Domain.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
2JTM
 
 | | Solution structure of Sso6901 from Sulfolobus solfataricus P2 | | Descriptor: | Putative uncharacterized protein | | Authors: | Feng, Y, Guo, L, Huang, L, Wang, J. | | Deposit date: | 2007-08-03 | | Release date: | 2008-04-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Biochemical and structural characterization of Cren7, a novel chromatin protein conserved among Crenarchaea
Nucleic Acids Res., 36, 2008
|
|
8HBA
 
 | |
8HB1
 
 | | Crystal structure of NAD-II riboswitch (two strands) with NMN | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, MAGNESIUM ION, RNA (30-MER), ... | | Authors: | Peng, X, Lilley, D.M.J, Huang, L. | | Deposit date: | 2022-10-27 | | Release date: | 2023-03-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Crystal structures of the NAD+-II riboswitch reveal two distinct ligand-binding pockets.
Nucleic Acids Res., 51, 2023
|
|
8HB8
 
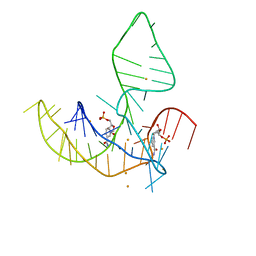 | |
8HB3
 
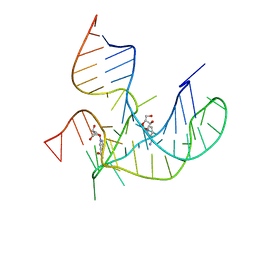 | | Crystal structure of NAD-II riboswitch (two strands) with NR | | Descriptor: | Nicotinamide riboside, RNA (31-MER), RNA (5'-R(*AP*GP*AP*GP*CP*GP*UP*UP*GP*CP*GP*UP*CP*CP*GP*AP*AP*AP*GP*UP*(CBV)P*GP*CP*C)-3') | | Authors: | Peng, X, Lilley, D.M.J, Huang, L. | | Deposit date: | 2022-10-27 | | Release date: | 2023-03-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Crystal structures of the NAD+-II riboswitch reveal two distinct ligand-binding pockets.
Nucleic Acids Res., 51, 2023
|
|
8I3Z
 
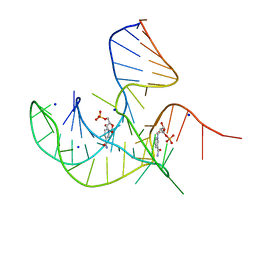 | | Crystal structure of NAD-II riboswitch (two strands) with NMN at 1.67 angstrom | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, RNA (31-MER), RNA (5'-R(*AP*GP*AP*GP*CP*GP*UP*UP*GP*CP*GP*UP*CP*CP*GP*AP*AP*AP*GP*UP*(CBV)P*GP*CP*C)-3'), ... | | Authors: | Peng, X, Lilley, D.M.J, Huang, L. | | Deposit date: | 2023-01-18 | | Release date: | 2023-03-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Crystal structures of the NAD+-II riboswitch reveal two distinct ligand-binding pockets.
Nucleic Acids Res., 51, 2023
|
|
6K8N
 
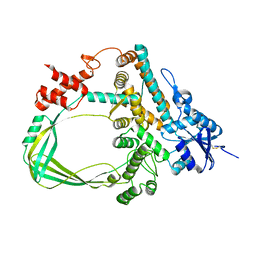 | | Crystal structure of the Sulfolobus solfataricus topoisomerase III | | Descriptor: | ZINC ION, topoisomerase III | | Authors: | Wang, H.Q, Zhang, J.H, Zheng, X, Zheng, Z.F, Dong, Y.H, Huang, L, Gong, Y. | | Deposit date: | 2019-06-13 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of the Sulfolobus solfataricus topoisomerase III reveal that its C-terminal novel zinc finger part is a unique decatenation domain
To Be Published
|
|
8ITS
 
 | |
6K8O
 
 | | Crystal structure of the Sulfolobus solfataricus topoisomerase III in complex with DNA | | Descriptor: | DNA (5'-D(*GP*CP*AP*AP*GP*GP*TP*C)-3'), ZINC ION, topoisomerase III | | Authors: | Wang, H.Q, Zhang, J.H, Zheng, X, Zheng, Z.F, Dong, Y.H, Huang, L, Gong, Y. | | Deposit date: | 2019-06-13 | | Release date: | 2020-06-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of the Sulfolobus solfataricus topoisomerase III reveal that its C-terminal novel zinc finger part is a unique decatenation domain
To Be Published
|
|
2KKE
 
 | | Solution NMR Structure of a dimeric protein of unknown function from Methanobacterium thermoautotrophicum, Northeast Structural Genomics Consortium Target TR5 | | Descriptor: | Uncharacterized protein | | Authors: | Swapna, G.V.T, Gunsalus, X, Huang, L, Xiao, K, Everett, J.K, Acton, T.B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-06-18 | | Release date: | 2009-07-14 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of a putative uncharacterized protein from Methanobacterium thermoautotrophicum, Northeast Structural Genomics Consortium Target:TR5
To be Published
|
|
7XI6
 
 | | Crystal structure of C56 from pSSVi | | Descriptor: | C56 | | Authors: | Zhang, Z.F, Ren, Y, Chen, Y.Y, Zhang, X.W, Dong, Y.H, Gong, Y, Cao, P, Huang, L. | | Deposit date: | 2022-04-12 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Characterization of the AbrB-like protein C56 conserved in a novel family of integrated genetic elements in Sulfolobales
To Be Published
|
|
7V9E
 
 | | Crystal structure of a methyl transferase ribozyme | | Descriptor: | BARIUM ION, GUANINE, RNA (68-MER), ... | | Authors: | Deng, J, Lilley, D.M.J, Huang, L. | | Deposit date: | 2021-08-25 | | Release date: | 2022-03-23 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and mechanism of a methyltransferase ribozyme.
Nat.Chem.Biol., 18, 2022
|
|
8JPX
 
 | | Cryo-EM structure of PfAgo-guide DNA-target DNA complex | | Descriptor: | Excess DNA, Guide DNA, MAGNESIUM ION, ... | | Authors: | Zhuang, L. | | Deposit date: | 2023-06-13 | | Release date: | 2024-01-31 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular mechanism for target recognition, dimerization, and activation of Pyrococcus furiosus Argonaute.
Mol.Cell, 84, 2024
|
|
8WD8
 
 | | Cryo-EM structure of TtdAgo-guide DNA-target DNA complex | | Descriptor: | Argonaute family protein, Guide DNA, MAGNESIUM ION, ... | | Authors: | Zhuang, L. | | Deposit date: | 2023-09-14 | | Release date: | 2024-01-31 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular mechanism for target recognition, dimerization, and activation of Pyrococcus furiosus Argonaute.
Mol.Cell, 84, 2024
|
|
3NO7
 
 | |
7YRD
 
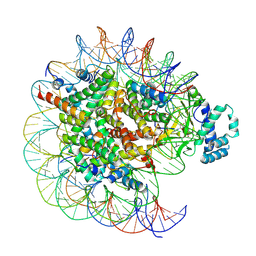 | | histone methyltransferase | | Descriptor: | DNA (146-MER), Histone H2A.Z, Histone H2B 1.1, ... | | Authors: | Li, H, Wang, W.Y. | | Deposit date: | 2022-08-09 | | Release date: | 2023-08-16 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insight into H4K20 methylation on H2A.Z-nucleosome by SUV420H1.
Mol.Cell, 83, 2023
|
|
7YRG
 
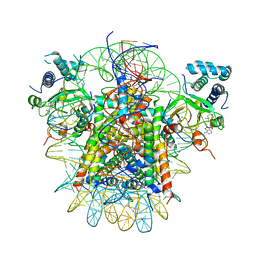 | | histone methyltransferase | | Descriptor: | DNA (146-MER), Histone H2A.Z, Histone H2B 1.1, ... | | Authors: | Li, H, Wang, W.Y. | | Deposit date: | 2022-08-09 | | Release date: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural insight into H4K20 methylation on H2A.Z-nucleosome by SUV420H1.
Mol.Cell, 83, 2023
|
|
6J3P
 
 | | Crystal structure of the human GCN5 bromodomain in complex with compound (R,R)-36n | | Descriptor: | 2-{[(3R,5R)-5-(2,3-dihydro-1,4-benzodioxin-6-yl)-1-methylpiperidin-3-yl]amino}-3-methyl-3,5-dihydro-4H-pyrrolo[3,2-d]pyrimidin-4-one, Histone acetyltransferase KAT2A | | Authors: | Huang, L.Y, Li, H, Niu, L, Wu, C.Y, Yu, Y.M, Li, L.L, Yang, S.Y. | | Deposit date: | 2019-01-05 | | Release date: | 2019-05-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.598 Å) | | Cite: | Discovery of Pyrrolo[3,2- d]pyrimidin-4-one Derivatives as a New Class of Potent and Cell-Active Inhibitors of P300/CBP-Associated Factor Bromodomain.
J.Med.Chem., 62, 2019
|
|
