6PZ4
 
 | | co-crystal structure of BACE with inhibitor AM-6494 | | Descriptor: | Beta-secretase 1, GLYCEROL, IODIDE ION, ... | | Authors: | Huang, X. | | Deposit date: | 2019-07-31 | | Release date: | 2019-10-23 | | Last modified: | 2020-03-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of AM-6494: A Potent and Orally Efficacious beta-Site Amyloid Precursor Protein Cleaving Enzyme 1 (BACE1) Inhibitor with in Vivo Selectivity over BACE2.
J.Med.Chem., 63, 2020
|
|
8UZA
 
 | | Cryo-EM structure of GeoCas9 in complex with sgRNA and target DNA | | Descriptor: | CRISPR-associated endonuclease Cas9, Non-target strand DNA, Target strand DNA, ... | | Authors: | Eggers, A.R, Soczek, K.M, Tuck, O.T, Doudna, J.A. | | Deposit date: | 2023-11-14 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Rapid DNA unwinding accelerates genome editing by engineered CRISPR-Cas9.
Cell, 187, 2024
|
|
8UZB
 
 | | Cryo-EM structure of iGeoCas9 in complex with sgRNA and target DNA | | Descriptor: | CRISPR-associated endonuclease Cas9, Non-target strand DNA, RNA (107-MER), ... | | Authors: | Eggers, A.R, Soczek, K.M, Tuck, O.T, Doudna, J.A. | | Deposit date: | 2023-11-14 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | Rapid DNA unwinding accelerates genome editing by engineered CRISPR-Cas9.
Cell, 187, 2024
|
|
8AXW
 
 | | The structure of mouse AsterC (GramD1c) with Ezetimibe | | Descriptor: | (3~{R},4~{S})-1-(4-fluorophenyl)-3-[(3~{S})-3-(4-fluorophenyl)-3-oxidanyl-propyl]-4-(4-hydroxyphenyl)azetidin-2-one, CHLORIDE ION, ETHANOL, ... | | Authors: | Fairall, L, Xiao, X, Burger, L, Tontonoz, P, Schwabe, J.W.R. | | Deposit date: | 2022-09-01 | | Release date: | 2023-09-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Aster-dependent nonvesicular transport facilitates dietary cholesterol uptake.
Science, 382, 2023
|
|
6M1K
 
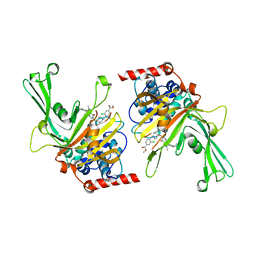 | | USP7 in complex with a novel inhibitor | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 7, methyl 4-[[4-[[3-[4-(aminomethyl)phenyl]-2-methyl-7-oxidanylidene-pyrazolo[4,3-d]pyrimidin-6-yl]methyl]-4-oxidanyl-piperidin-1-yl]methyl]-3-chloranyl-benzoate | | Authors: | Liu, S.J, Zhou, X.Y, Li, M.L, Sun, H.B, Wen, X.A. | | Deposit date: | 2020-02-26 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.255 Å) | | Cite: | N-benzylpiperidinol derivatives as novel USP7 inhibitors: Structure-activity relationships and X-ray crystallographic studies.
Eur.J.Med.Chem., 199, 2020
|
|
4DI2
 
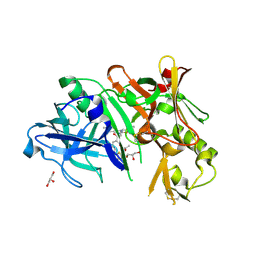 | | Crystal structure of BACE1 in complex with hydroxyethylamine inhibitor 37 | | Descriptor: | (2R)-N-{(2S,3R)-4-{[(4'S)-6'-(2,2-dimethylpropyl)-3',4'-dihydrospiro[cyclobutane-1,2'-pyrano[2,3-b]pyridin]-4'-yl]amino}-3-hydroxy-1-[3-(1,3-thiazol-2-yl)phenyl]butan-2-yl}-2-methoxypropanamide, Beta-secretase 1, GLYCEROL | | Authors: | Whittington, D.A, Long, A.M. | | Deposit date: | 2012-01-30 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design and synthesis of potent, orally efficacious hydroxyethylamine derived beta-site amyloid precursor protein cleaving enzyme (BACE1) inhibitors.
J.Med.Chem., 55, 2012
|
|
4FRJ
 
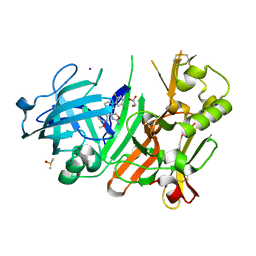 | | Crystal structure of BACE1 in complex with aminooxazoline xanthene 9l | | Descriptor: | (4S)-2'-(5-chloro-2-fluorophenyl)-7'-methoxyspiro[1,3-oxazole-4,9'-xanthen]-2-amine, Beta-secretase 1, DIMETHYL SULFOXIDE, ... | | Authors: | Whittington, D.A, Long, A.M. | | Deposit date: | 2012-06-26 | | Release date: | 2012-09-12 | | Last modified: | 2012-12-12 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure- and Property-Based Design of Aminooxazoline Xanthenes as Selective, Orally Efficacious, and CNS Penetrable BACE Inhibitors for the Treatment of Alzheimer's Disease.
J.Med.Chem., 55, 2012
|
|
5X8I
 
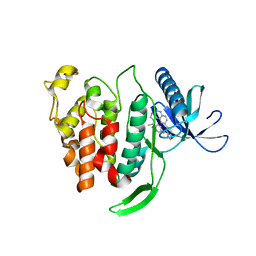 | | Crystal structure of human CLK1 in complex with compound 25 | | Descriptor: | 5-[1-[(1S)-1-(4-fluorophenyl)ethyl]-[1,2,3]triazolo[4,5-c]quinolin-8-yl]-1,3-benzoxazole, Dual specificity protein kinase CLK1 | | Authors: | Sun, Q.Z, Lin, G.F, Li, L.L, Jin, X.T, Huang, L.Y, Zhang, G, Wei, Y.Q, Lu, G.W, Yang, S.Y. | | Deposit date: | 2017-03-02 | | Release date: | 2017-08-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Discovery of Potent and Selective Inhibitors of Cdc2-Like Kinase 1 (CLK1) as a New Class of Autophagy Inducers
J. Med. Chem., 60, 2017
|
|
6VB9
 
 | | Covalent adduct of cis-2,3-epoxysuccinic acid with Isocitrate Lyase-1 from Mycobacterium tuberculosis | | Descriptor: | ACETIC ACID, DI(HYDROXYETHYL)ETHER, Isocitrate lyase, ... | | Authors: | Krieger, I.V, Pham, T.V, Meek, T.D, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2019-12-18 | | Release date: | 2020-12-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.881 Å) | | Cite: | Covalent Inactivation of Mycobacterium tuberculosis Isocitrate Lyase by cis -2,3-Epoxy-Succinic Acid.
Acs Chem.Biol., 16, 2021
|
|
4FRK
 
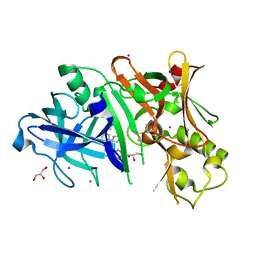 | | Crystal structure of BACE1 in complex with aminooxazoline xanthene 11a | | Descriptor: | (4S)-2'-(2-methylpropoxy)-7'-(pyrimidin-5-yl)spiro[1,3-oxazole-4,9'-xanthen]-2-amine, Beta-secretase 1, GLYCEROL, ... | | Authors: | Whittington, D.A, Long, A.M. | | Deposit date: | 2012-06-26 | | Release date: | 2012-09-12 | | Last modified: | 2012-12-12 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure- and Property-Based Design of Aminooxazoline Xanthenes as Selective, Orally Efficacious, and CNS Penetrable BACE Inhibitors for the Treatment of Alzheimer's Disease.
J.Med.Chem., 55, 2012
|
|
4FRI
 
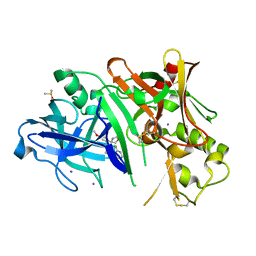 | | Crystal structure of BACE1 in complex with biarylspiro aminooxazoline 6 | | Descriptor: | (4R)-4-[3-(2-fluoropyridin-3-yl)phenyl]-4-(4-methoxyphenyl)-4,5-dihydro-1,3-oxazol-2-amine, Beta-secretase 1, DIMETHYL SULFOXIDE, ... | | Authors: | Whittington, D.A, Long, A.M. | | Deposit date: | 2012-06-26 | | Release date: | 2012-09-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure- and Property-Based Design of Aminooxazoline Xanthenes as Selective, Orally Efficacious, and CNS Penetrable BACE Inhibitors for the Treatment of Alzheimer's Disease.
J.Med.Chem., 55, 2012
|
|
4FS4
 
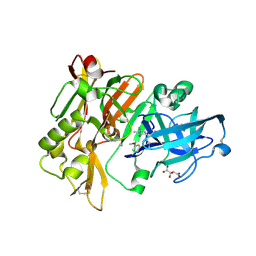 | | Structure of BACE Bound to (S)-4-(3'-methoxy-[1,1'-biphenyl]-3-yl)-1,4-dimethyl-6-oxotetrahydropyrimidin-2(1H)-iminium | | Descriptor: | (6S)-2-amino-6-(3'-methoxybiphenyl-3-yl)-3,6-dimethyl-5,6-dihydropyrimidin-4(3H)-one, Beta-secretase 1, L(+)-TARTARIC ACID | | Authors: | Strickland, C, Stamford, A. | | Deposit date: | 2012-06-26 | | Release date: | 2012-10-10 | | Last modified: | 2014-07-23 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | A Potent and Orally Efficacious, Hydroxyethylamine-Based Inhibitor of beta-Secretase.
ACS Med Chem Lett, 3, 2012
|
|
1U5S
 
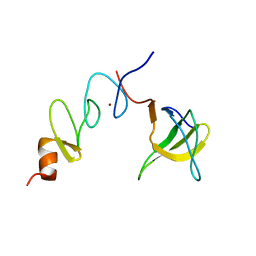 | | NMR structure of the complex between Nck-2 SH3 domain and PINCH-1 LIM4 domain | | Descriptor: | Cytoplasmic protein NCK2, PINCH protein, ZINC ION | | Authors: | Vaynberg, J, Fukuda, T, Vinogradova, O, Velyvis, A, Ng, L, Wu, C, Qin, J. | | Deposit date: | 2004-07-28 | | Release date: | 2005-04-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of an ultraweak protein-protein complex and its crucial role in regulation of cell morphology and motility.
Mol.Cell, 17, 2005
|
|
2A4R
 
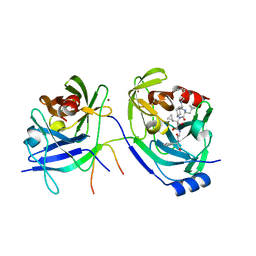 | | HCV NS3 Protease Domain with a Ketoamide Inhibitor Covalently bound. | | Descriptor: | NS3 protease/helicase, Ns4a peptide, ZINC ION, ... | | Authors: | Bogen, S, Saksena, A.K, Arasappan, A, Gu, H, Njoroge, F.G, Girijavallabhan, V, Pichardo, J, Butkiewicz, N, Prongay, A, Madison, V. | | Deposit date: | 2005-06-29 | | Release date: | 2006-07-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Hepatitis C Virus NS3-4A serine protease inhibitors: Use of a P2-P1 cyclopropyl alanine combination for improved potency.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
3DP2
 
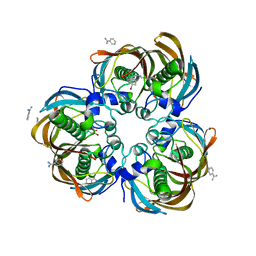 | | Crystal structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase (FabZ) from Helicobacter pylori in complex with compound 3j | | Descriptor: | (3R)-hydroxymyristoyl-acyl carrier protein dehydratase, 4-bromo-N'-[(1E)-(3,5-dibromo-2,4-dihydroxyphenyl)methylidene]benzohydrazide, BENZAMIDINE, ... | | Authors: | Zhang, L, He, L, Liu, X, Liu, H, Shen, X, Jiang, H. | | Deposit date: | 2008-07-07 | | Release date: | 2009-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovering potent inhibitors against the beta-hydroxyacyl-acyl carrier protein dehydratase (FabZ) of Helicobacter pylori: structure-based design, synthesis, bioassay, and crystal structure determination.
J.Med.Chem., 52, 2009
|
|
3DOZ
 
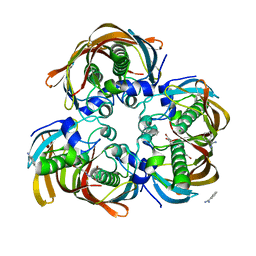 | | Crystal structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase (FabZ) from Helicobacter pylori in complex with compound 3k | | Descriptor: | (3R)-hydroxymyristoyl-acyl carrier protein dehydratase, 3-bromo-N'-[(1E)-(3,5-dibromo-2,4-dihydroxyphenyl)methylidene]benzohydrazide, BENZAMIDINE, ... | | Authors: | Zhang, L, He, L, Liu, X, Liu, H, Shen, X, Jiang, H. | | Deposit date: | 2008-07-07 | | Release date: | 2009-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovering potent inhibitors against the beta-hydroxyacyl-acyl carrier protein dehydratase (FabZ) of Helicobacter pylori: structure-based design, synthesis, bioassay, and crystal structure determination.
J.Med.Chem., 52, 2009
|
|
3DOY
 
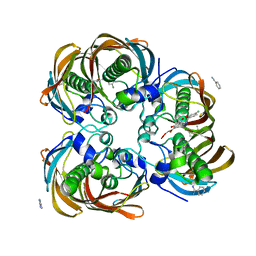 | | Crystal structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase (FabZ) from Helicobacter pylori in complex with compound 3i | | Descriptor: | (3R)-hydroxymyristoyl-acyl carrier protein dehydratase, 4-chloro-N'-[(1E)-(3,5-dibromo-2,4-dihydroxyphenyl)methylidene]benzohydrazide, BENZAMIDINE, ... | | Authors: | Zhang, L, He, L, Liu, X, Liu, H, Shen, X, Jiang, H. | | Deposit date: | 2008-07-07 | | Release date: | 2009-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovering potent inhibitors against the beta-hydroxyacyl-acyl carrier protein dehydratase (FabZ) of Helicobacter pylori: structure-based design, synthesis, bioassay, and crystal structure determination.
J.Med.Chem., 52, 2009
|
|
3DP3
 
 | | Crystal structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase (FabZ) from Helicobacter pylori in complex with compound 3q | | Descriptor: | (3R)-hydroxymyristoyl-acyl carrier protein dehydratase, 4-tert-butyl-N'-[(1E)-(3,5-dibromo-2,4-dihydroxyphenyl)methylidene]benzohydrazide, BENZAMIDINE, ... | | Authors: | Zhang, L, He, L, Liu, X, Liu, H, Shen, X, Jiang, H. | | Deposit date: | 2008-07-07 | | Release date: | 2009-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovering potent inhibitors against the beta-hydroxyacyl-acyl carrier protein dehydratase (FabZ) of Helicobacter pylori: structure-based design, synthesis, bioassay, and crystal structure determination.
J.Med.Chem., 52, 2009
|
|
3DP1
 
 | | Crystal structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase (FabZ) from Helicobacter pylori in complex with compound 3n | | Descriptor: | (3R)-hydroxymyristoyl-acyl carrier protein dehydratase, BENZAMIDINE, CHLORIDE ION, ... | | Authors: | Zhang, L, He, L, Liu, X, Liu, H, Shen, X, Jiang, H. | | Deposit date: | 2008-07-07 | | Release date: | 2009-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovering potent inhibitors against the beta-hydroxyacyl-acyl carrier protein dehydratase (FabZ) of Helicobacter pylori: structure-based design, synthesis, bioassay, and crystal structure determination.
J.Med.Chem., 52, 2009
|
|
3DP0
 
 | | Crystal structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase (FabZ) from Helicobacter pylori in complex with compound 3m | | Descriptor: | (3R)-hydroxymyristoyl-acyl carrier protein dehydratase, BENZAMIDINE, CHLORIDE ION, ... | | Authors: | Zhang, L, He, L, Liu, X, Liu, H, Shen, X, Jiang, H. | | Deposit date: | 2008-07-07 | | Release date: | 2009-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovering potent inhibitors against the beta-hydroxyacyl-acyl carrier protein dehydratase (FabZ) of Helicobacter pylori: structure-based design, synthesis, bioassay, and crystal structure determination.
J.Med.Chem., 52, 2009
|
|
7CWO
 
 | | SARS-CoV-2 spike protein RBD and P17 fab complex | | Descriptor: | Spike glycoprotein, heavy chain of P17 Fab, light chain of P17 Fab | | Authors: | Wang, X, Wang, N. | | Deposit date: | 2020-08-29 | | Release date: | 2020-12-16 | | Last modified: | 2021-01-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Rational development of a human antibody cocktail that deploys multiple functions to confer Pan-SARS-CoVs protection.
Cell Res., 31, 2021
|
|
7CWL
 
 | |
2O8M
 
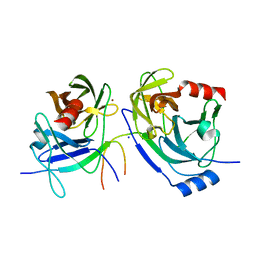 | | Crystal structure of the S139A mutant of Hepatitis C Virus NS3/4A protease | | Descriptor: | Protease, SODIUM ION, ZINC ION | | Authors: | Fischmann, T.O, Prongay, A.J, Madison, V.M, Yao, N. | | Deposit date: | 2006-12-12 | | Release date: | 2007-10-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3- [2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]- 6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034) II. Key steps in structure-based optimization
J.Med.Chem., 50, 2007
|
|
7CWM
 
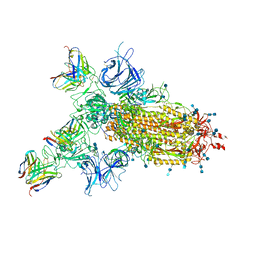 | |
7CWN
 
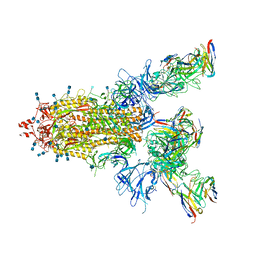 | | P17-H014 Fab cocktail in complex with SARS-CoV-2 spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Wang, N, Wang, X. | | Deposit date: | 2020-08-29 | | Release date: | 2020-12-16 | | Last modified: | 2021-01-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Rational development of a human antibody cocktail that deploys multiple functions to confer Pan-SARS-CoVs protection.
Cell Res., 31, 2021
|
|
