1RNJ
 
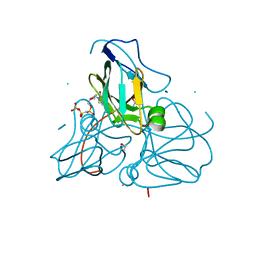 | | Crystal structure of inactive mutant dUTPase complexed with substrate analogue imido-dUTP | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Deoxyuridine 5'-triphosphate nucleotidohydrolase, ... | | Authors: | Barabas, O, Pongracz, V, Kovari, J, Wilmanns, M, Vertessy, B.G. | | Deposit date: | 2003-12-01 | | Release date: | 2004-09-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Insights into the Catalytic Mechanism of Phosphate Ester Hydrolysis by dUTPase.
J.Biol.Chem., 279, 2004
|
|
2R15
 
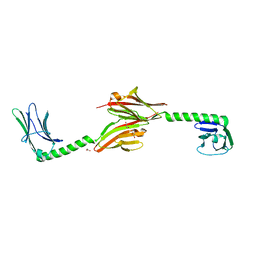 | |
1RUW
 
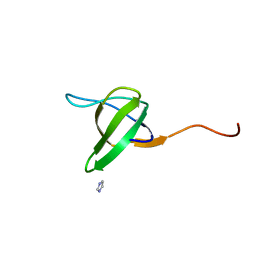 | | Crystal structure of the SH3 domain from S. cerevisiae Myo3 | | Descriptor: | IMIDAZOLE, Myosin-3 isoform | | Authors: | Kursula, P, Kursula, I, Lehmann, F, Song, Y.H, Wilmanns, M. | | Deposit date: | 2003-12-12 | | Release date: | 2005-03-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the SH3 domain from S. cerevisiae Myo3
To be Published
|
|
1RN8
 
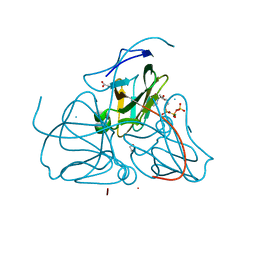 | | Crystal structure of dUTPase complexed with substrate analogue imido-dUTP | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Barabas, O, Pongracz, V, Kovari, J, Wilmanns, M, Vertessy, B.G. | | Deposit date: | 2003-12-01 | | Release date: | 2004-09-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural Insights into the Catalytic Mechanism of Phosphate Ester Hydrolysis by dUTPase.
J.Biol.Chem., 279, 2004
|
|
1SEH
 
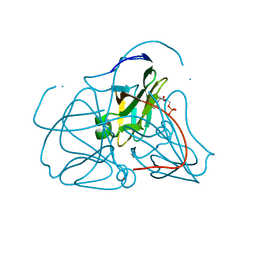 | | Crystal structure of E. coli dUTPase complexed with the product dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Deoxyuridine 5'-triphosphate nucleotidohydrolase | | Authors: | Barabas, O, Kovari, J, Pongracz, V, Wilmanns, M, Vertessy, B.G. | | Deposit date: | 2004-02-17 | | Release date: | 2004-09-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structural Insights into the Catalytic Mechanism of Phosphate Ester Hydrolysis by dUTPase
J.Biol.Chem., 279, 2004
|
|
1TG0
 
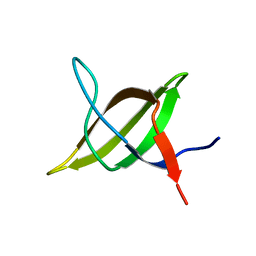 | |
2V1R
 
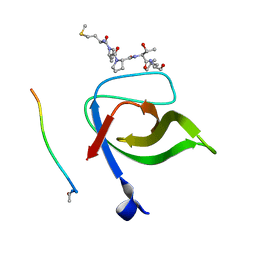 | | Yeast Pex13 SH3 domain complexed with a peptide from Pex14 at 2.1 A resolution | | Descriptor: | PEROXISOMAL MEMBRANE PROTEIN PAS20, PEX14 | | Authors: | Kursula, I, Kursula, P, Lehmann, F, Zou, P, Song, Y.H, Wilmanns, M. | | Deposit date: | 2007-05-29 | | Release date: | 2008-06-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Genomics of Yeast SH3 Domains
To be Published
|
|
1SSH
 
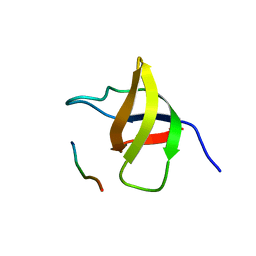 | | Crystal structure of the SH3 domain from a S. cerevisiae hypothetical 40.4 kDa protein in complex with a peptide | | Descriptor: | 12-mer peptide from Cytoskeleton assembly control protein SLA1, Hypothetical 40.4 kDa protein in PES4-HIS2 intergenic region | | Authors: | Kursula, P, Kursula, I, Lehmann, F, Song, Y.-H, Wilmanns, M. | | Deposit date: | 2004-03-24 | | Release date: | 2005-04-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Yeast SH3 domain structural genomics
To be Published
|
|
1SYL
 
 | | Crystal structure of inactive mutant dUTPase complexed with substrate dUTP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DEOXYURIDINE-5'-TRIPHOSPHATE, Deoxyuridine 5'-triphosphate nucleotidohydrolase, ... | | Authors: | Barabas, O, Kovari, J, Pongracz, V, Wilmanns, M, Vertessy, B.G. | | Deposit date: | 2004-04-01 | | Release date: | 2004-09-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Insights into the Catalytic Mechanism of Phosphate Ester Hydrolysis by dUTPase
J.Biol.Chem., 279, 2004
|
|
2VKN
 
 | | YEAST SHO1 SH3 DOMAIN COMPLEXED WITH A PEPTIDE FROM PBS2 | | Descriptor: | MAP KINASE KINASE PBS2, PROTEIN SSU81, SULFATE ION | | Authors: | Kursula, P, Kursula, I, Song, Y.H, Paraskevopoulos, K, Wilmanns, M. | | Deposit date: | 2007-12-20 | | Release date: | 2008-02-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Genomics of Yeast SH3 Domains
To be Published
|
|
2V1Q
 
 | | Atomic-resolution structure of the yeast Sla1 SH3 domain 3 | | Descriptor: | CHLORIDE ION, CYTOSKELETON ASSEMBLY CONTROL PROTEIN SLA1, PLATINUM (II) ION, ... | | Authors: | Kursula, I, Kursula, P, Zou, P, Lehmann, F, Song, Y.H, Wilmanns, M. | | Deposit date: | 2007-05-29 | | Release date: | 2008-06-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural Genomics of Yeast SH3 Domains
To be Published
|
|
2J8O
 
 | | Structure of the immunoglobulin tandem repeat of titin A168-A169 | | Descriptor: | GLYCEROL, TITIN | | Authors: | Mueller, S, Lange, S, Kursula, I, Gautel, M, Wilmanns, M. | | Deposit date: | 2006-10-26 | | Release date: | 2007-08-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Rigid Conformation of an Immunoglobulin Domain Tandem Repeat in the A-Band of the Elastic Muscle Protein Titin
J.Mol.Biol., 371, 2007
|
|
2J8H
 
 | | Structure of the immunoglobulin tandem repeat A168-A169 of titin | | Descriptor: | GLYCEROL, TITIN | | Authors: | Mueller, S, Lange, S, Kursula, I, Gautel, M, Wilmanns, M. | | Deposit date: | 2006-10-25 | | Release date: | 2007-08-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Rigid Conformation of an Immunoglobulin Domain Tandem Repeat in the A-Band of the Elastic Muscle Protein Titin
J.Mol.Biol., 371, 2007
|
|
2WL8
 
 | | X-ray crystal structure of Pex19p | | Descriptor: | PEROXISOMAL BIOGENESIS FACTOR 19 | | Authors: | Schueller, N, Holton, S.J, Stanley, W.A, Song, Y.H, Konarev, P, Roessle, M, Erdmann, R, Schliebs, W, Wilmanns, M. | | Deposit date: | 2009-06-22 | | Release date: | 2010-06-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The Peroxisomal Receptor Pex19P Forms a Helical Mpts Recognition Domain.
Embo J., 29, 2010
|
|
4I8A
 
 | | Alanine-glyoxylate aminotransferase variant S187F | | Descriptor: | GLYCEROL, Serine-pyruvate aminotransferase | | Authors: | Fodor, K, Oppici, E, Williams, C, Cellini, B, Wilmanns, M. | | Deposit date: | 2012-12-03 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the S187F variant of human liver alanine: Aminotransferase associated with primary hyperoxaluria type I and its functional implications.
Proteins, 81, 2013
|
|
3ZL4
 
 | | Antibody structural organization: Role of kappa - lambda chain constant domain switch in catalytic functionality | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, A17 ANTIBODY FAB FRAGMENT HEAVY CHAIN, A17 ANTIBODY FAB FRAGMENT LAMBDA LIGHT CHAIN | | Authors: | Chatziefthimiou, S.D, Ponomarenko, N.A, Kurkova, I.N, Smirnov, A.V, Smirnov, I.V, Lamzin, V.S, Gabibov, A.G, Wilmanns, M. | | Deposit date: | 2013-01-28 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Role of Kappa>Lambda Light-Chain Constant-Domain Switch in the Structure and Functionality of A17 Reactibody
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4KXK
 
 | | Alanine-glyoxylate aminotransferase variant K390A/K391A in complex with the TPR domain of human Pex5p | | Descriptor: | BETA-MERCAPTOETHANOL, Peroxisomal targeting signal 1 receptor, SULFATE ION, ... | | Authors: | Fodor, K, Lou, Y, Wilmanns, M. | | Deposit date: | 2013-05-27 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Ligand-Induced Compaction of the PEX5 Receptor-Binding Cavity Impacts Protein Import Efficiency into Peroxisomes.
Traffic, 16, 2015
|
|
4KYO
 
 | | Alanine-glyoxylate aminotransferase variant K390A in complex with the TPR domain of human Pex5p | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, Peroxisomal targeting signal 1 receptor, ... | | Authors: | Fodor, K, Lou, Y, Wilmanns, M. | | Deposit date: | 2013-05-29 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Ligand-Induced Compaction of the PEX5 Receptor-Binding Cavity Impacts Protein Import Efficiency into Peroxisomes.
Traffic, 16, 2015
|
|
2W85
 
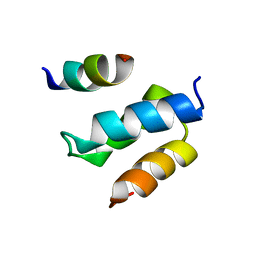 | | Structure of Pex14 in complex with Pex19 | | Descriptor: | PEROXIN-19, PEROXISOMAL MEMBRANE ANCHOR PROTEIN PEX14 | | Authors: | Neufeld, C, Filipp, F.V, Simon, B, Neuhaus, A, Schueller, N, David, C, Kooshapur, H, Madl, T, Erdmann, R, Schliebs, W, Wilmanns, M, Sattler, M. | | Deposit date: | 2009-01-09 | | Release date: | 2009-02-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Competitive Interactions of Pex14 with the Import Receptors Pex5 and Pex19.
Embo J., 28, 2009
|
|
2YQ4
 
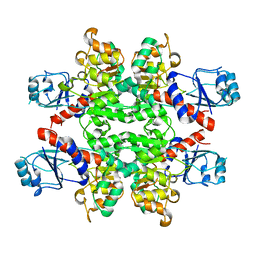 | |
2Y9M
 
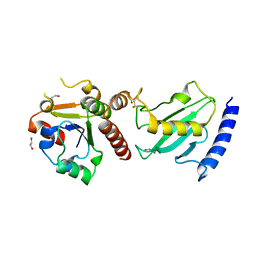 | | Pex4p-Pex22p structure | | Descriptor: | 1,2-ETHANEDIOL, PEROXISOME ASSEMBLY PROTEIN 22, UBIQUITIN-CONJUGATING ENZYME E2-21 KDA | | Authors: | Williams, C, van den Berg, M, Panjikar, S, Distel, B, Wilmanns, M. | | Deposit date: | 2011-02-15 | | Release date: | 2011-10-26 | | Last modified: | 2017-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Insights Into Ubiquitin-Conjugating Enzyme/ Co-Activator Interactions from the Structure of the Pex4P:Pex22P Complex.
Embo J., 31, 2011
|
|
2YQ5
 
 | | Crystal Structure of D-isomer specific 2-hydroxyacid dehydrogenase from Lactobacillus delbrueckii ssp. bulgaricus: NAD complexed form | | Descriptor: | D-ISOMER SPECIFIC 2-HYDROXYACID DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Holton, S.J, Anandhakrishnan, M, Geerlof, A, Wilmanns, M. | | Deposit date: | 2012-11-05 | | Release date: | 2012-11-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural Characterization of D-Isomer Specific 2-Hydroxyacid Dehydrogenase from Lactobacillus Delbrueckii Ssp. Bulgaricus
J.Struct.Biol., 181, 2013
|
|
2Y9P
 
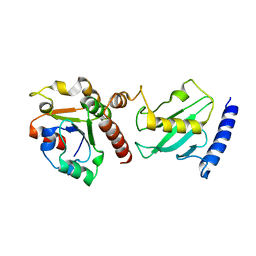 | | Pex4p-Pex22p mutant II structure | | Descriptor: | PEROXISOME ASSEMBLY PROTEIN 22, UBIQUITIN-CONJUGATING ENZYME E2-21 KDA | | Authors: | Williams, C, van den Berg, M, Panjikar, S, Distel, B, Wilmanns, M. | | Deposit date: | 2011-02-16 | | Release date: | 2012-02-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Pex4P-Pex22P Structure
To be Published
|
|
1UUE
 
 | | a-SPECTRIN SH3 DOMAIN (V44T, D48G MUTANT) | | Descriptor: | SPECTRIN ALPHA CHAIN | | Authors: | Vega, M.C, Fernandez, A, Wilmanns, M, Serrano, L. | | Deposit date: | 2003-12-18 | | Release date: | 2004-02-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Solvation in Protein Folding Analysis: Combination of Theoretical and Experimental Approaches
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
2Y25
 
 | |
