7RCY
 
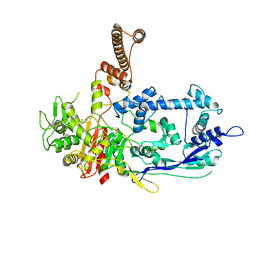 | | Crystal structure of C. difficile penicillin-binding protein 2 in complex with ceftobiprole | | Descriptor: | (2R)-2-[(1R)-1-{[(2Z)-2-(5-amino-1,2,4-thiadiazol-3-yl)-2-(hydroxyimino)acetyl]amino}-2-oxoethyl]-5-({2-oxo-1-[(3R)-pyr rolidin-3-yl]-2,5-dihydro-1H-pyrrol-3-yl}methyl)-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Penicillin-binding protein, ZINC ION | | Authors: | Sacco, M, Chen, Y. | | Deposit date: | 2021-07-08 | | Release date: | 2022-03-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A unique class of Zn 2+ -binding serine-based PBPs underlies cephalosporin resistance and sporogenesis in Clostridioides difficile.
Nat Commun, 13, 2022
|
|
7RCX
 
 | |
5IVB
 
 | |
5IVC
 
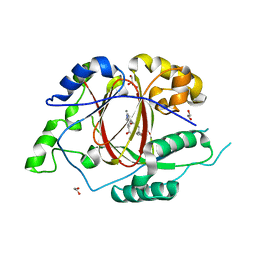 | | Linked KDM5A Jmj Domain Bound to the Inhibitor N3 (4'-[(2-phenylethyl)carbamoyl][2,2'-bipyridine]-4-carboxylic acid) | | Descriptor: | 1,2-ETHANEDIOL, 4'-[(2-phenylethyl)carbamoyl][2,2'-bipyridine]-4-carboxylic acid, GLYCEROL, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2016-03-20 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.573 Å) | | Cite: | Structural Basis for KDM5A Histone Lysine Demethylase Inhibition by Diverse Compounds.
Cell Chem Biol, 23, 2016
|
|
5IWF
 
 | |
5IVF
 
 | | Linked KDM5A Jmj Domain Bound to the Inhibitor N10 8-(1-methyl-1H-imidazol-4-yl)-2-(4,4,4-trifluorobutoxy)pyrido[3,4-d]pyrimidin-4-ol | | Descriptor: | 8-(1-methyl-1H-imidazol-4-yl)-2-(4,4,4-trifluorobutoxy)pyrido[3,4-d]pyrimidin-4-ol, Lysine-specific demethylase 5A, MANGANESE (II) ION | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2016-03-20 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.683 Å) | | Cite: | Structural Basis for KDM5A Histone Lysine Demethylase Inhibition by Diverse Compounds.
Cell Chem Biol, 23, 2016
|
|
5IVV
 
 | |
5ZJI
 
 | | Structure of photosystem I supercomplex with light-harvesting complexes I and II | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Pan, X.W, Ma, J, Su, X.D, Cao, P, Liu, Z.F, Zhang, X.Z, Li, M. | | Deposit date: | 2018-03-20 | | Release date: | 2018-06-20 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of the maize photosystem I supercomplex with light-harvesting complexes I and II.
Science, 360, 2018
|
|
8ISJ
 
 | | Pr conformer of Arabidopsis thaliana phytochrome A - AtphyA-Pr | | Descriptor: | 3-[5-[[(3~{R},4~{R})-3-ethyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, Phytochrome A | | Authors: | Zhang, Y, Ma, C, Zhao, J, Gao, N, Wang, J. | | Deposit date: | 2023-03-20 | | Release date: | 2023-08-09 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into plant phytochrome A as a highly sensitized photoreceptor.
Cell Res., 33, 2023
|
|
8ISI
 
 | | Photochromobilin-free form of Arabidopsis thaliana phytochrome A - apo-AtphyA | | Descriptor: | Phytochrome A | | Authors: | Zhang, Y, Ma, C, Zhao, J, Gao, N, Wang, J. | | Deposit date: | 2023-03-20 | | Release date: | 2023-08-09 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Structural insights into plant phytochrome A as a highly sensitized photoreceptor.
Cell Res., 33, 2023
|
|
6OD5
 
 | | Human TCF4 C-terminal bHLH domain in Complex with 12-bp Oligonucleotide Containing E-box Sequence with 5-carboxylcytosines | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA (5'-D(*AP*(1CC)P*GP*CP*AP*CP*GP*TP*GP*(1CC)P*G)-3'), ... | | Authors: | Horton, J.R, Cheng, X, Yang, J. | | Deposit date: | 2019-03-25 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for preferential binding of human TCF4 to DNA containing 5-carboxylcytosine.
Nucleic Acids Res., 47, 2019
|
|
8H83
 
 | | Crystal structure of a IsPETase variant V22 from Ideonella sakaiensis | | Descriptor: | Poly(ethylene terephthalate) hydrolase | | Authors: | Wei, H.L, Gao, S.F, Li, Q, Han, X, Gao, J, Liu, W.D. | | Deposit date: | 2022-10-21 | | Release date: | 2024-04-24 | | Last modified: | 2025-02-12 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | beta-sheet Engineering of IsPETase for PET Depolymerization
Engineering (Beijing), 2024
|
|
7BRM
 
 | | Architecture of curli complex | | Descriptor: | Curli production assembly/transport protein CsgG, csgf | | Authors: | Zhang, M, Shi, H. | | Deposit date: | 2020-03-29 | | Release date: | 2020-07-15 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of the nonameric CsgG-CsgF complex and its implications for controlling curli biogenesis in Enterobacteriaceae.
Plos Biol., 18, 2020
|
|
5IVJ
 
 | | Linked KDM5A Jmj Domain Bound to the Inhibitor N11 [3-({1-[2-(4,4-difluoropiperidin-1-yl)ethyl]-5-fluoro-1H-indazol-3-yl}amino)pyridine-4-carboxylic acid] | | Descriptor: | 3-({1-[2-(4,4-difluoropiperidin-1-yl)ethyl]-5-fluoro-1H-indazol-3-yl}amino)pyridine-4-carboxylic acid, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2016-03-21 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.569 Å) | | Cite: | Structural Basis for KDM5A Histone Lysine Demethylase Inhibition by Diverse Compounds.
Cell Chem Biol, 23, 2016
|
|
5IVY
 
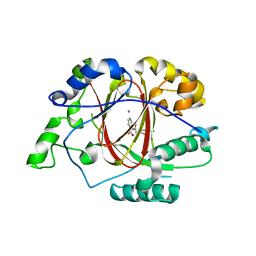 | |
5IVE
 
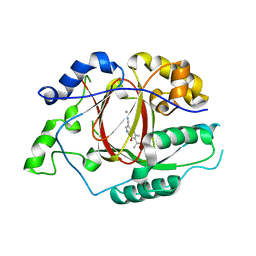 | | Linked KDM5A Jmj Domain Bound to the Inhibitor N8 ( 5-methyl-7-oxo-6-(propan-2-yl)-4,7-dihydropyrazolo[1,5-a]pyrimidine-3-carbonitrile) | | Descriptor: | 5-methyl-7-oxo-6-(propan-2-yl)-4,7-dihydropyrazolo[1,5-a]pyrimidine-3-carbonitrile, Lysine-specific demethylase 5A, MANGANESE (II) ION | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2016-03-20 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.783 Å) | | Cite: | Structural Basis for KDM5A Histone Lysine Demethylase Inhibition by Diverse Compounds.
Cell Chem Biol, 23, 2016
|
|
5IW0
 
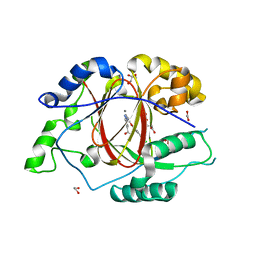 | |
8P5Y
 
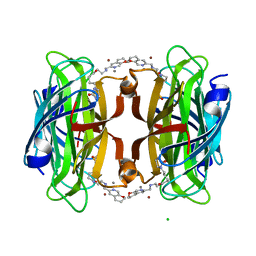 | | Artificial transfer hydrogenase with a Mn-12 cofactor and Streptavidin S112Y-K121M mutant | | Descriptor: | 5-[(3~{a}~{S},4~{S},6~{a}~{R})-2-oxidanylidene-1,3,3~{a},4,6,6~{a}-hexahydrothieno[3,4-d]imidazol-4-yl]-~{N}-[2-(3,4-dihydro-2~{H}-pyrano[2,3-c]pyridin-6-ylmethylamino)ethyl]pentanamide, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Lau, K, Wang, W, Pojer, F, Larabi, A. | | Deposit date: | 2023-05-24 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Manganese Transfer Hydrogenases Based on the Biotin-Streptavidin Technology.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8P5Z
 
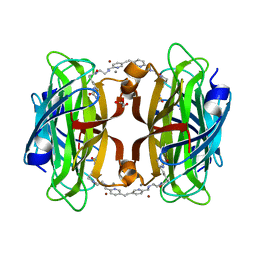 | | Artificial transfer hydrogenase with a Mn-5 cofactor and Streptavidin S112Y-K121M mutant | | Descriptor: | 5-[(3~{a}~{S},4~{S},6~{a}~{R})-2-oxidanylidene-1,3,3~{a},4,6,6~{a}-hexahydrothieno[3,4-d]imidazol-4-yl]-~{N}-[2-[(5-methylpyridin-2-yl)methylamino]ethyl]pentanamide, BROMIDE ION, GLYCEROL, ... | | Authors: | Lau, K, Wang, W, Pojer, F, Larabi, A. | | Deposit date: | 2023-05-24 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Manganese Transfer Hydrogenases Based on the Biotin-Streptavidin Technology.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
6WK1
 
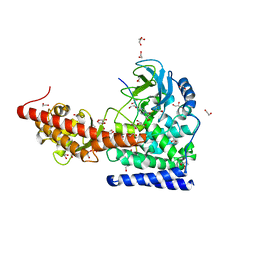 | | SETD3 in Complex with an Actin Peptide with His73 Replaced with Methionine | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Actin, ... | | Authors: | Dai, S, Horton, J.R, Cheng, X. | | Deposit date: | 2020-04-15 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Characterization of SETD3 methyltransferase-mediated protein methionine methylation.
J.Biol.Chem., 295, 2020
|
|
6XFN
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with UAW243 | | Descriptor: | 3C-like proteinase, GLYCEROL, UAW243 | | Authors: | Sacco, M, Ma, C, Wang, J, Chen, Y. | | Deposit date: | 2020-06-15 | | Release date: | 2020-06-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and inhibition of the SARS-CoV-2 main protease reveal strategy for developing dual inhibitors against M pro and cathepsin L.
Sci Adv, 6, 2020
|
|
6XBI
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with inhibitor UAW248 | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Sacco, M, Ma, C, Wang, J, Chen, Y. | | Deposit date: | 2020-06-06 | | Release date: | 2020-06-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and inhibition of the SARS-CoV-2 main protease reveal strategy for developing dual inhibitors against M pro and cathepsin L.
Sci Adv, 6, 2020
|
|
8C4A
 
 | |
5V3J
 
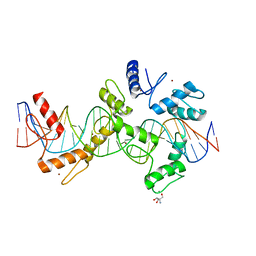 | | mouseZFP568-ZnF1-10 in complex with DNA | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DNA (26-MER), MAGNESIUM ION, ... | | Authors: | Patel, A, Cheng, X. | | Deposit date: | 2017-03-07 | | Release date: | 2018-03-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.064 Å) | | Cite: | DNA Conformation Induces Adaptable Binding by Tandem Zinc Finger Proteins.
Cell, 173, 2018
|
|
5V3M
 
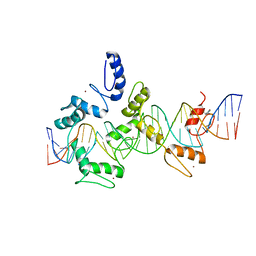 | | mouseZFP568-ZnF1-11 in complex with DNA | | Descriptor: | DNA (28-MER), ZINC ION, Zinc finger protein 568 | | Authors: | Patel, A, Cheng, X. | | Deposit date: | 2017-03-07 | | Release date: | 2018-03-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.091 Å) | | Cite: | DNA Conformation Induces Adaptable Binding by Tandem Zinc Finger Proteins.
Cell, 173, 2018
|
|
