3K0G
 
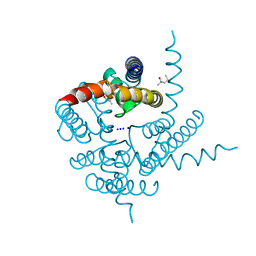 | | Crystal Structure of CNG mimicking NaK mutant, NaK-ETPP, Na+ complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Potassium channel protein NaK, SODIUM ION | | Authors: | Jiang, Y, Derebe, M.G. | | Deposit date: | 2009-09-24 | | Release date: | 2011-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural studies of ion permeation and Ca2+ blockage of a bacterial channel mimicking the cyclic nucleotide-gated channel pore.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
6ILO
 
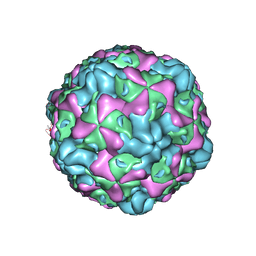 | | Cryo-EM structure of empty Echovirus 6 particle at PH 7.4 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Gao, G.F, Liu, S, Zhao, X, Peng, R. | | Deposit date: | 2018-10-19 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Human Neonatal Fc Receptor Is the Cellular Uncoating Receptor for Enterovirus B.
Cell, 177, 2019
|
|
5GOZ
 
 | |
6ILL
 
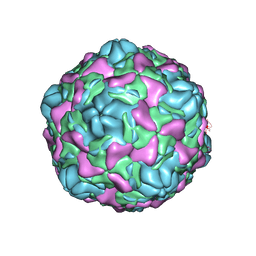 | | Cryo-EM structure of Echovirus 6 complexed with its uncoating receptor FcRn at PH 5.5 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Gao, G.F, Liu, S, Zhao, X, Peng, R. | | Deposit date: | 2018-10-19 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Human Neonatal Fc Receptor Is the Cellular Uncoating Receptor for Enterovirus B.
Cell, 177, 2019
|
|
6ILJ
 
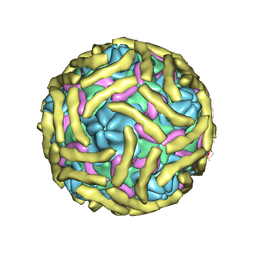 | | Cryo-EM structure of Echovirus 6 complexed with its attachment receptor CD55 at PH 5.5 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Gao, G.F, Liu, S, Zhao, X, Peng, R. | | Deposit date: | 2018-10-18 | | Release date: | 2019-05-15 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Human Neonatal Fc Receptor Is the Cellular Uncoating Receptor for Enterovirus B.
Cell, 177, 2019
|
|
6SZ1
 
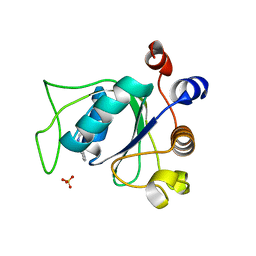 | | Crystal structure of YTHDC1 with fragment 2 (DHU_DC1_140) | | Descriptor: | SULFATE ION, YTH domain-containing protein 1, ~{N}-methylquinazolin-4-amine | | Authors: | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | Deposit date: | 2019-10-01 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Selectively Disrupting m6A-Dependent Protein-RNA Interactions with Fragments.
Acs Chem.Biol., 15, 2020
|
|
6T05
 
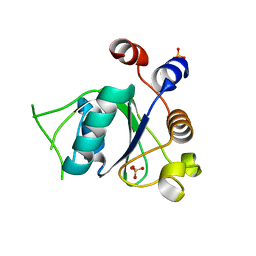 | | Crystal structure of YTHDC1 with fragment 18 (DHU_DC1_048) | | Descriptor: | 2-(phenylmethyl)imidazolidine, SULFATE ION, YTHDC1 | | Authors: | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | Deposit date: | 2019-10-02 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Selectively Disrupting m6A-Dependent Protein-RNA Interactions with Fragments.
Acs Chem.Biol., 15, 2020
|
|
6T0D
 
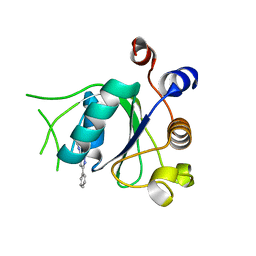 | | Crystal structure of YTHDC1 with fragment 27 (DHU_DC1_256) | | Descriptor: | SULFATE ION, YTHDC1, ~{N}-methyl-3-phenyl-1~{H}-pyrazole-5-carboxamide | | Authors: | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | Deposit date: | 2019-10-02 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Selectively Disrupting m6A-Dependent Protein-RNA Interactions with Fragments.
Acs Chem.Biol., 15, 2020
|
|
4TXR
 
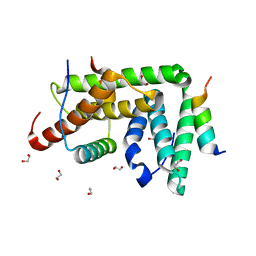 | |
6T10
 
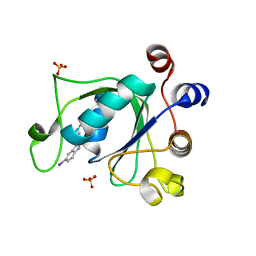 | | Crystal structure of YTHDC1 with fragment 28 (DHU_DC1_176) | | Descriptor: | 5-iodanyl-~{N}-methyl-1~{H}-indazole-3-carboxamide, SULFATE ION, YTHDC1 | | Authors: | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | Deposit date: | 2019-10-03 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Selectively Disrupting m6A-Dependent Protein-RNA Interactions with Fragments.
Acs Chem.Biol., 15, 2020
|
|
6SZL
 
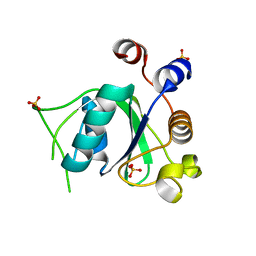 | | Crystal structure of YTHDC1 with fragment 7 (DHU_DC1_021) | | Descriptor: | 6-phenyl-1~{H}-pyrimidine-2,4-dione, SULFATE ION, YTH domain-containing protein 1 | | Authors: | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | Deposit date: | 2019-10-02 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Selectively Disrupting m6A-Dependent Protein-RNA Interactions with Fragments.
Acs Chem.Biol., 15, 2020
|
|
6T09
 
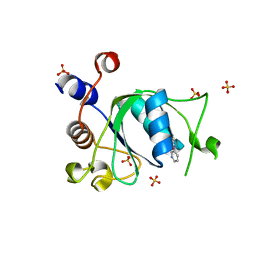 | | Crystal structure of YTHDC1 with fragment 24 (PSI_DC1_003) | | Descriptor: | SULFATE ION, YTHDC1, ~{N}-pyridin-3-ylethanamide | | Authors: | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | Deposit date: | 2019-10-02 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Selectively Disrupting m6A-Dependent Protein-RNA Interactions with Fragments.
Acs Chem.Biol., 15, 2020
|
|
4TXQ
 
 | | Crystal structure of LIP5 N-terminal domain complexed with CHMP1B MIM | | Descriptor: | Charged multivesicular body protein 1b, Vacuolar protein sorting-associated protein VTA1 homolog | | Authors: | Vild, C.J, Xu, Z. | | Deposit date: | 2014-07-04 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | A Novel Mechanism of Regulating the ATPase VPS4 by Its Cofactor LIP5 and the Endosomal Sorting Complex Required for Transport (ESCRT)-III Protein CHMP5.
J.Biol.Chem., 290, 2015
|
|
6U0D
 
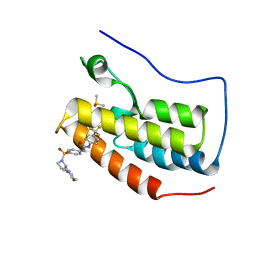 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor ZL0590 | | Descriptor: | Bromodomain-containing protein 4, DIMETHYL SULFOXIDE, N-[4-({(2S)-2-[(morpholin-4-yl)methyl]pyrrolidin-1-yl}sulfonyl)phenyl]-N'-[4-(trifluoromethyl)phenyl]urea | | Authors: | Leonard, P.G, Joseph, S. | | Deposit date: | 2019-08-14 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Discovery, X-ray Crystallography, and Anti-inflammatory Activity of Bromodomain-containing Protein 4 (BRD4) BD1 Inhibitors Targeting a Distinct New Binding Site
J. Med. Chem., 2022
|
|
7E3J
 
 | |
3SX0
 
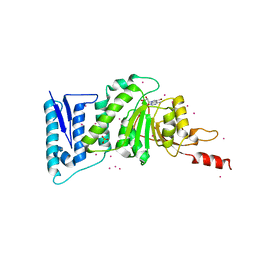 | | Crystal structure of Dot1l in complex with a brominated SAH analog | | Descriptor: | (2S)-2-amino-4-({[(2S,3S,4R,5R)-5-(4-amino-5-bromo-7H-pyrrolo[2,3-d]pyrimidin-7-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl}sulfanyl)butanoic acid (non-preferred name), Histone-lysine N-methyltransferase, H3 lysine-79 specific, ... | | Authors: | Yu, W, Tempel, W, Smil, D, Schapira, M, Li, Y, Vedadi, M, Nguyen, K.T, Wernimont, A.K, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-07-14 | | Release date: | 2011-07-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Bromo-deaza-SAH: a potent and selective DOT1L inhibitor.
Bioorg. Med. Chem., 21, 2013
|
|
8FIS
 
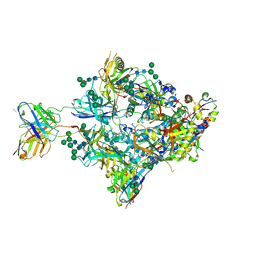 | | Structure of Bispecific CAP256V2LS-J3 Fab in complex with BG505 DS-SOSIP.664 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Gorman, J, Kwong, P.D. | | Deposit date: | 2022-12-16 | | Release date: | 2023-02-01 | | Last modified: | 2023-02-15 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Bispecific antibody CAP256.J3LS targets V2-apex and CD4-binding sites with high breadth and potency.
Mabs, 15, 2023
|
|
4TXP
 
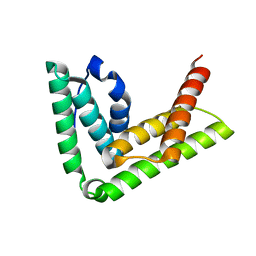 | | Crystal structure of LIP5 N-terminal domain | | Descriptor: | Vacuolar protein sorting-associated protein VTA1 homolog | | Authors: | Vild, C.J, Xu, Z. | | Deposit date: | 2014-07-04 | | Release date: | 2015-02-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | A Novel Mechanism of Regulating the ATPase VPS4 by Its Cofactor LIP5 and the Endosomal Sorting Complex Required for Transport (ESCRT)-III Protein CHMP5.
J.Biol.Chem., 290, 2015
|
|
6UKB
 
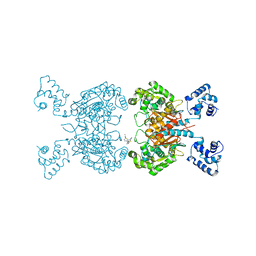 | | Crystal structure of human GAC in complex with inhibitor UPGL00012 | | Descriptor: | Glutaminase kidney isoform, mitochondrial, N-[5-(4-{[5-(acetylamino)-1,3,4-thiadiazol-2-yl]oxy}piperidin-1-yl)-1,3,4-thiadiazol-2-yl]acetamide | | Authors: | Huang, Q, Cerione, R.A. | | Deposit date: | 2019-10-04 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of human GAC in complex with inhibitor UPGL00020
To Be Published
|
|
7E0B
 
 | |
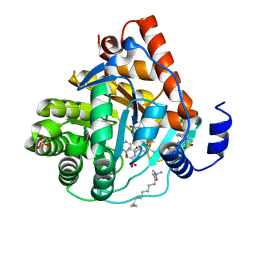
3KVL
 
 | |
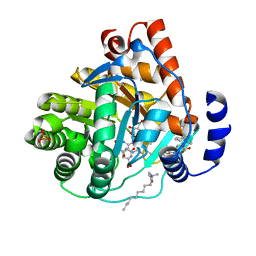
3KVJ
 
 | |
3KVK
 
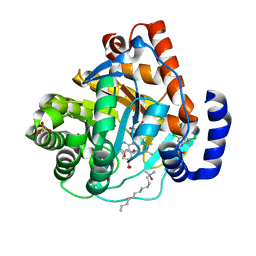 | | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with amino-benzoic acid inhibitor 641 at 2.05A resolution | | Descriptor: | (4S)-2,6-DIOXOHEXAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, 2-{[(3,5-dichlorophenyl)carbamoyl]amino}benzoic acid, Dihydroorotate dehydrogenase, ... | | Authors: | McLean, L, Zhang, Y. | | Deposit date: | 2009-11-30 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of novel inhibitors for DHODH via virtual screening and X-ray crystallographic structures.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
5V0L
 
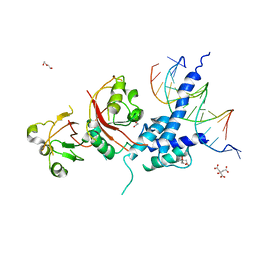 | | Crystal structure of the AHR-ARNT heterodimer in complex with the DRE | | Descriptor: | Aryl hydrocarbon receptor, Aryl hydrocarbon receptor nuclear translocator, CITRIC ACID, ... | | Authors: | Seok, S.-H, Lee, W, Jiang, L, Bradfield, C.A, Xing, Y. | | Deposit date: | 2017-02-28 | | Release date: | 2017-04-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structural hierarchy controlling dimerization and target DNA recognition in the AHR transcriptional complex.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7D0A
 
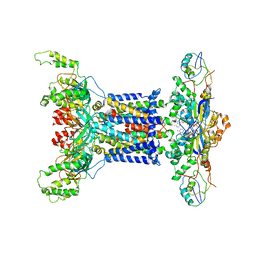 | | Acinetobacter MlaFEDB complex in ADP-vanadate trapped Vclose conformation | | Descriptor: | ABC transporter ATP-binding protein, ADENOSINE-5'-DIPHOSPHATE, Anti-sigma factor antagonist, ... | | Authors: | Zhang, Y.Y, Fan, Q.X, Chi, X.M, Zhou, Q, Li, Y.Y. | | Deposit date: | 2020-09-09 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structures of Acinetobacter baumannii glycerophospholipid transporter.
Cell Discov, 6, 2020
|
|
