6QN6
 
 | |
8ZUF
 
 | | Cryo-EM structure of P.nat ACE2 mutant in complex with MOW15-22 RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Tang, J, Deng, Z. | | Deposit date: | 2024-06-09 | | Release date: | 2025-02-12 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Multiple independent acquisitions of ACE2 usage in MERS-related coronaviruses.
Cell, 188, 2025
|
|
5GR8
 
 | | Crystal structure of PEPR1-AtPEP1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Elicitor peptide 1, ... | | Authors: | Chai, J.J, Tang, J. | | Deposit date: | 2016-08-08 | | Release date: | 2016-12-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.587 Å) | | Cite: | Structural basis for recognition of an endogenous peptide by the plant receptor kinase PEPR1
Cell Res., 25, 2015
|
|
4BF1
 
 | | Three dimensional structure of human carbonic anhydrase II in complex with 5-(1-naphthalen-1-yl-1,2,3-triazol-4-yl)thiophene-2-sulfonamide | | Descriptor: | 5-(1-naphthalen-1-yl-1,2,3-triazol-4-yl)thiophene-2-sulfonamide, CARBONIC ANHYDRASE 2, SODIUM ION, ... | | Authors: | Tars, K, Leitans, J, Zalubovskis, R. | | Deposit date: | 2013-03-13 | | Release date: | 2014-01-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | 5-Substituted-(1,2,3-Triazol-4-Yl)Thiophene-2-Sulfonamides Strongly Inhibit Human Carbonic Anhydrases I, II, Ix and Xii: Solution and X-Ray Crystallographic Studies.
Bioorg.Med.Chem., 21, 2013
|
|
4BF6
 
 | | Three dimensional structure of human carbonic anhydrase II in complex with 5-(1-(3-Cyanophenyl)-1H-1,2,3-triazol-4-yl)thiophene-2- sulfonamide | | Descriptor: | 5-[1-(3-cyanophenyl)-1,2,3-triazol-4-yl]thiophene-2-sulfonamide, CARBONIC ANHYDRASE 2, GLYCEROL, ... | | Authors: | Tars, K, Leitans, J, Zalubovskis, R. | | Deposit date: | 2013-03-15 | | Release date: | 2014-01-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | 5-Substituted-(1,2,3-Triazol-4-Yl)Thiophene-2-Sulfonamides Strongly Inhibit Human Carbonic Anhydrases I, II, Ix and Xii: Solution and X-Ray Crystallographic Studies.
Bioorg.Med.Chem., 21, 2013
|
|
5AMD
 
 | |
5AML
 
 | |
5AMG
 
 | |
2OYV
 
 | | Neurotensin in DPC micelles | | Descriptor: | neurotensin | | Authors: | Monti, J.P, Coutant, J, Curmi, P.A. | | Deposit date: | 2007-02-23 | | Release date: | 2007-05-08 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of Neurotensin in Membrane-Mimetic Environments: Molecular Basis for Neurotensin Receptor Recognition.
Biochemistry, 46, 2007
|
|
2OYW
 
 | | Neurotensin in TFE:H2O (80:20) | | Descriptor: | neurotensin | | Authors: | Monti, J.P, Coutant, J, Curmi, P.A. | | Deposit date: | 2007-02-23 | | Release date: | 2007-05-08 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of Neurotensin in Membrane-Mimetic Environments: Molecular Basis for Neurotensin Receptor Recognition.
Biochemistry, 46, 2007
|
|
8YQ8
 
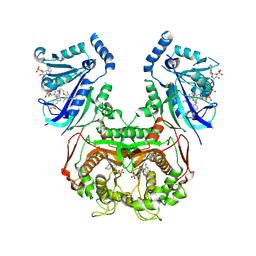 | | Quadruple mutant (N51I+C59R+S108N+I164L) Plasmodium falciparum dihydrofolate reductase-thymidylate synthase (PfDHFR-TS V1/S) complexed with FB8, NADPH and dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 4-[4-[4-[2,4-bis(azanyl)-6-ethyl-pyrimidin-5-yl]oxybutoxy]phenyl]benzoic acid, Bifunctional dihydrofolate reductase-thymidylate synthase, ... | | Authors: | Vanichtanankul, J, Vitsupakorn, D, Saeyang, T, Arwon, U, Hoarau, M, Decharuangsilp, S, Kamchonwongpaisan, S, Yuthavong, Y. | | Deposit date: | 2024-03-19 | | Release date: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Novel flexible biphenyl Pf DHFR inhibitors with improved antimalarial activity.
Rsc Med Chem, 15, 2024
|
|
8YQ9
 
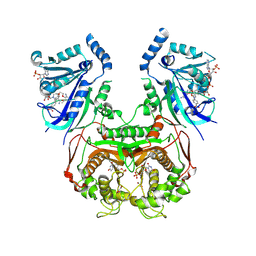 | | Quadruple mutant (N51I+C59R+S108N+I164L) Plasmodium falciparum dihydrofolate reductase-thymidylate synthase (PfDHFR-TS V1/S) complexed with FB6, NADPH and dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 4-[4-[3-[2,4-bis(azanyl)-6-ethyl-pyrimidin-5-yl]oxypropoxy]phenyl]benzoic acid, Bifunctional dihydrofolate reductase-thymidylate synthase, ... | | Authors: | Vanichtanankul, J, Vitsupakorn, D, Saeyang, T, Arwon, U, Hoarau, M, Decharuangsilp, S, Kamchonwongpaisan, S, Yuthavong, Y. | | Deposit date: | 2024-03-19 | | Release date: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Novel flexible biphenyl Pf DHFR inhibitors with improved antimalarial activity.
Rsc Med Chem, 15, 2024
|
|
7E24
 
 | |
7E28
 
 | |
7E3X
 
 | | Crystal structure of SDR family NAD(P)-dependent oxidoreductase from exiguobacterium | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Oxidoreductase | | Authors: | Chen, L, Tang, J, Yuan, S, Zhang, F, Chen, S. | | Deposit date: | 2021-02-09 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structure-guided evolution of a ketoreductase forefficient and stereoselective bioreduction of bulkyalpha-aminobeta-keto esters
Catalysis Science And Technology, 2021
|
|
7F3Y
 
 | | Wild-type Plasmodium falciparum dihydrofolate reductase-thymidylate synthase (PfDHFR-TS) complexed with methotrexate (MTX), NADPH and dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, Bifunctional dihydrofolate reductase-thymidylate synthase, GLYCEROL, ... | | Authors: | Vanichtanankul, J, Tanramluk, D, Yuvaniyama, J, Yuthavong, Y. | | Deposit date: | 2021-06-17 | | Release date: | 2021-09-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.252 Å) | | Cite: | MANORAA: A machine learning platform to guide protein-ligand design by anchors and influential distances.
Structure, 30, 2022
|
|
7F3Z
 
 | | Double mutant Plasmodium falciparum dihydrofolate reductase-thymidylate synthase (PfDHFR-TS-K1, C59R+S108N) complexed with Trimethoprim (TOP), NADPH and dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, Bifunctional dihydrofolate reductase-thymidylate synthase, GLYCEROL, ... | | Authors: | Vanichtanankul, J, Tanramluk, D, Chitnumsub, P, Yuvaniyama, J, Yuthavong, Y. | | Deposit date: | 2021-06-17 | | Release date: | 2021-09-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | MANORAA: A machine learning platform to guide protein-ligand design by anchors and influential distances.
Structure, 30, 2022
|
|
6A7E
 
 | | Human dihydrofolate reductase complexed with NADPH and BT2 | | Descriptor: | 5-(4-{3-[(2,4-diamino-6-ethylpyrimidin-5-yl)oxy]propoxy}phenyl)-6-ethylpyrimidine-2,4-diamine, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Vanichtanankul, J, Tarnchompoo, B, Chitnumsub, P, Kamchonwongpaisan, S, Yuthavong, Y. | | Deposit date: | 2018-07-02 | | Release date: | 2019-04-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Hybrid Inhibitors of Malarial Dihydrofolate Reductase with Dual Binding Modes That Can Forestall Resistance.
ACS Med Chem Lett, 9, 2018
|
|
4PT2
 
 | |
6A7C
 
 | | Human dihydrofolate reductase complexed with NADPH and BT1 | | Descriptor: | 5-(3-{3-[(2,4-diamino-6-ethylpyrimidin-5-yl)oxy]propoxy}phenyl)-6-ethylpyrimidine-2,4-diamine, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Vanichtanankul, J, Tarnchompoo, B, Chitnumsub, P, Kamchonwongpaisan, S, Yuthavong, Y. | | Deposit date: | 2018-07-02 | | Release date: | 2019-04-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Hybrid Inhibitors of Malarial Dihydrofolate Reductase with Dual Binding Modes That Can Forestall Resistance.
ACS Med Chem Lett, 9, 2018
|
|
6KP7
 
 | |
6KP2
 
 | |
6KOT
 
 | |
7CTW
 
 | | Wild-type Plasmodium falciparum dihydrofolate reductase-thymidylate synthase (PfDHFR-TS) complexed with fragment 820, NADPH, dUMP | | Descriptor: | 1-(2-methylsulfanylphenyl)piperazine, Bifunctional dihydrofolate reductase-thymidylate synthase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Vanichtanankul, J, Vitsupakorn, D. | | Deposit date: | 2020-08-20 | | Release date: | 2021-04-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Discovery of new non-pyrimidine scaffolds as Plasmodium falciparum DHFR inhibitors by fragment-based screening.
J Enzyme Inhib Med Chem, 36, 2021
|
|
7CTY
 
 | | Wild type plasmodium falciparum dihydrofolate reductase-thymidylate synthase (PfDHFR-TS), fragment 263, NADP+, dUMP | | Descriptor: | Bifunctional dihydrofolate reductase-thymidylate synthase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PHOSPHATE ION, ... | | Authors: | Vanichtanankul, J, Vitsupakorn, D. | | Deposit date: | 2020-08-20 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of new non-pyrimidine scaffolds as Plasmodium falciparum DHFR inhibitors by fragment-based screening.
J Enzyme Inhib Med Chem, 36, 2021
|
|
