8GTM
 
 | | Corticotropin-releasing hormone receptor 1(CRF1R) bound with BMK-C203 by XFEL | | Descriptor: | 7-(4-bromanyl-2,6-dimethoxy-phenyl)-4,8-dimethyl-~{N},~{N}-bis[4,4,4-tris(fluoranyl)butyl]-1$l^{4},3,5,9-tetrazabicyclo[4.3.0]nona-1(6),2,4,8-tetraen-2-amine, Endolysin, Isoform CRF-R2 of Corticotropin-releasing factor receptor 1 | | Authors: | Cho, H.S, Kim, H. | | Deposit date: | 2022-09-08 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-based drug discovery of a corticotropin-releasing hormone receptor 1 antagonist using an X-ray free-electron laser.
Exp.Mol.Med., 55, 2023
|
|
7WUC
 
 | |
4LW4
 
 | |
4LW2
 
 | |
6J43
 
 | | Proteinase K determined by PAL-XFEL | | Descriptor: | CALCIUM ION, Proteinase K | | Authors: | Lee, S.J, Park, J. | | Deposit date: | 2019-01-07 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Application of a high-throughput microcrystal delivery system to serial femtosecond crystallography.
J.Appl.Crystallogr., 53, 2020
|
|
7SVM
 
 | | DPP8 IN COMPLEX WITH LIGAND ICeD-2 | | Descriptor: | (2S)-2-amino-1-(1,3-dihydro-2H-isoindol-2-yl)-2-[(1r,4S)-4-(pyrrolidin-1-yl)cyclohexyl]ethan-1-one, Dipeptidyl peptidase 8, trimethylamine oxide | | Authors: | Lammens, A, Hollenstein, K, Klein, D.J. | | Deposit date: | 2021-11-19 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | A Phenotypic Screen Identifies Potent DPP9 Inhibitors Capable of Killing HIV-1 Infected Cells.
Acs Chem.Biol., 17, 2022
|
|
7SVO
 
 | | DPP8 IN COMPLEX WITH LIGAND ICeD-1 | | Descriptor: | (2S,4S)-1-[(2S)-2-amino-2-cyclohexylacetyl]-4-fluoropyrrolidine-2-carbonitrile, Dipeptidyl peptidase 8, trimethylamine oxide | | Authors: | Lammens, A, Hollenstein, K, Klein, D.J. | | Deposit date: | 2021-11-19 | | Release date: | 2022-10-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | A Phenotypic Screen Identifies Potent DPP9 Inhibitors Capable of Killing HIV-1 Infected Cells.
Acs Chem.Biol., 17, 2022
|
|
7SVN
 
 | | DPP9 IN COMPLEX WITH LIGAND ICeD-1 | | Descriptor: | (2S,4S)-1-[(2S)-2-amino-2-cyclohexylacetyl]-4-fluoropyrrolidine-2-carbonitrile, Dipeptidyl peptidase 9 | | Authors: | Lammens, A, Hollenstein, K, Klein, D.J. | | Deposit date: | 2021-11-19 | | Release date: | 2022-10-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | A Phenotypic Screen Identifies Potent DPP9 Inhibitors Capable of Killing HIV-1 Infected Cells.
Acs Chem.Biol., 17, 2022
|
|
7SVL
 
 | | DPP9 IN COMPLEX WITH LIGAND ICeD-2 | | Descriptor: | (2S)-2-amino-1-(1,3-dihydro-2H-isoindol-2-yl)-2-[(1r,4S)-4-(pyrrolidin-1-yl)cyclohexyl]ethan-1-one, Dipeptidyl peptidase 9 | | Authors: | Lammens, A, Hollenstein, K, Klein, D.J. | | Deposit date: | 2021-11-19 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | A Phenotypic Screen Identifies Potent DPP9 Inhibitors Capable of Killing HIV-1 Infected Cells.
Acs Chem.Biol., 17, 2022
|
|
7ML5
 
 | | Structure of the Starch Branching Enzyme I (BEI) complexed with maltododecaose from Oryza sativa L | | Descriptor: | Isoform 2 of 1,4-alpha-glucan-branching enzyme, chloroplastic/amyloplastic, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Nayebi Gavgani, H, Fawaz, R, Geiger, J.H. | | Deposit date: | 2021-04-27 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A structural explanation for the mechanism and specificity of plant branching enzymes I and IIb.
J.Biol.Chem., 298, 2021
|
|
5KUT
 
 | | hMiro2 C-terminal GTPase domain, GDP-bound | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Mitochondrial Rho GTPase 2 | | Authors: | Klosowiak, J.L, Focia, P.J, Rice, S.E, Freymann, D.M. | | Deposit date: | 2016-07-13 | | Release date: | 2016-09-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.693 Å) | | Cite: | Structural insights into Parkin substrate lysine targeting from minimal Miro substrates.
Sci Rep, 6, 2016
|
|
5KSZ
 
 | | hMiro EF hand and cGTPase domains in the GMPPCP-bound state | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Mitochondrial Rho GTPase 1, ... | | Authors: | Klosowiak, J.L, Focia, P.J, Rice, S.E, Freymann, D.M. | | Deposit date: | 2016-07-10 | | Release date: | 2016-09-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into Parkin substrate lysine targeting from minimal Miro substrates.
Sci Rep, 6, 2016
|
|
5KSY
 
 | | hMiro1 C-domain GDP Complex P41212 Crystal Form | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Mitochondrial Rho GTPase 1 | | Authors: | Klosowiak, J.L, Focia, P.J, Rice, S.E, Freymann, D.M. | | Deposit date: | 2016-07-10 | | Release date: | 2016-09-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.482 Å) | | Cite: | Structural insights into Parkin substrate lysine targeting from minimal Miro substrates.
Sci Rep, 6, 2016
|
|
5KTY
 
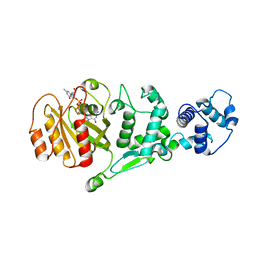 | | hMiro EF hand and cGTPase domains, GDP and Ca2+ bound state | | Descriptor: | CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Klosowiak, J.L, Focia, P.J, Rice, S.E, Freymann, D.M. | | Deposit date: | 2016-07-12 | | Release date: | 2016-09-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.522 Å) | | Cite: | Structural insights into Parkin substrate lysine targeting from minimal Miro substrates.
Sci Rep, 6, 2016
|
|
5KSP
 
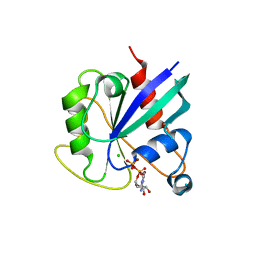 | | hMiro1 C-domain GDP Complex C2221 Crystal Form | | Descriptor: | CHLORIDE ION, GUANOSINE-5'-DIPHOSPHATE, Mitochondrial Rho GTPase 1 | | Authors: | Klosowiak, J.L, Focia, P.J, Rice, S.E, Freymann, D.M. | | Deposit date: | 2016-07-08 | | Release date: | 2016-09-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.162 Å) | | Cite: | Structural insights into Parkin substrate lysine targeting from minimal Miro substrates.
Sci Rep, 6, 2016
|
|
5KSO
 
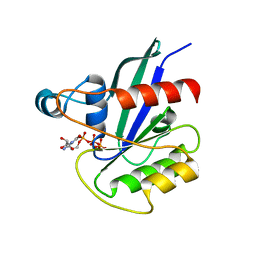 | | hMiro1 C-domain GDP-Pi Complex P3121 Crystal Form | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Mitochondrial Rho GTPase 1, PHOSPHATE ION | | Authors: | Klosowiak, J.L, Focia, P.J, Rice, S.E, Freymann, D.M. | | Deposit date: | 2016-07-08 | | Release date: | 2016-09-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural insights into Parkin substrate lysine targeting from minimal Miro substrates.
Sci Rep, 6, 2016
|
|
5KU1
 
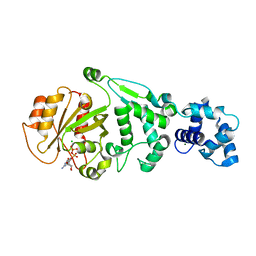 | | hMiro1 EF hand and cGTPase domains in the GDP-bound state | | Descriptor: | CHLORIDE ION, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Klosowiak, J.L, Focia, P.J, Rice, S.E, Freymann, D.M. | | Deposit date: | 2016-07-12 | | Release date: | 2016-09-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Structural insights into Parkin substrate lysine targeting from minimal Miro substrates.
Sci Rep, 6, 2016
|
|
6L18
 
 | |
4WO1
 
 | | Crystal structure of the DAP12 transmembrane domain in lipid cubic phase | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CALCIUM ION, TYRO protein tyrosine kinase-binding protein | | Authors: | Call, M.J, Call, M.E, Knoblich, K. | | Deposit date: | 2014-10-15 | | Release date: | 2015-06-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Transmembrane Complexes of DAP12 Crystallized in Lipid Membranes Provide Insights into Control of Oligomerization in Immunoreceptor Assembly.
Cell Rep, 11, 2015
|
|
4WOL
 
 | | Crystal Structure of the DAP12 transmembrane domain in lipidic cubic phase | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, POTASSIUM ION, TYRO protein tyrosine kinase-binding protein | | Authors: | Call, M.J, Call, M.E, Knoblich, K. | | Deposit date: | 2014-10-16 | | Release date: | 2015-06-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Transmembrane Complexes of DAP12 Crystallized in Lipid Membranes Provide Insights into Control of Oligomerization in Immunoreceptor Assembly.
Cell Rep, 11, 2015
|
|
5Z2E
 
 | |
7JL4
 
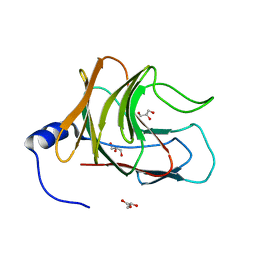 | | Crystal structure of TRIM65 PSpry domain | | Descriptor: | GLYCEROL, Tripartite motif-containing protein 65 | | Authors: | Kato, K, Ahmad, S, Hur, S. | | Deposit date: | 2020-07-29 | | Release date: | 2020-12-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural analysis of RIG-I-like receptors reveals ancient rules of engagement between diverse RNA helicases and TRIM ubiquitin ligases.
Mol.Cell, 81, 2021
|
|
5Z2F
 
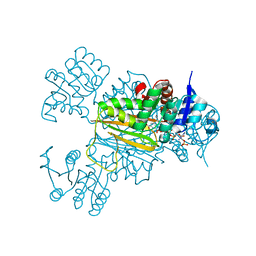 | | NADPH/PDA bound Dihydrodipicolinate reductase from Paenisporosarcina sp. TG-14 | | Descriptor: | Dihydrodipicolinate reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PYRIDINE-2,6-DICARBOXYLIC ACID | | Authors: | Lee, J.H, Lee, C.W, Park, S. | | Deposit date: | 2018-01-02 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of dihydrodipicolinate reductase (PaDHDPR) from Paenisporosarcina sp. TG-14: structural basis for NADPH preference as a cofactor
Sci Rep, 8, 2018
|
|
7JL3
 
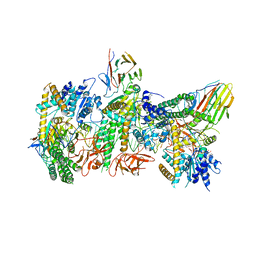 | | Cryo-EM structure of RIG-I:dsRNA filament in complex with RIPLET PrySpry domain (trimer) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Antiviral innate immune response receptor RIG-I, E3 ubiquitin-protein ligase RNF135, ... | | Authors: | Kato, K, Ahmad, S, Hur, S. | | Deposit date: | 2020-07-29 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural analysis of RIG-I-like receptors reveals ancient rules of engagement between diverse RNA helicases and TRIM ubiquitin ligases.
Mol.Cell, 81, 2021
|
|
7JL0
 
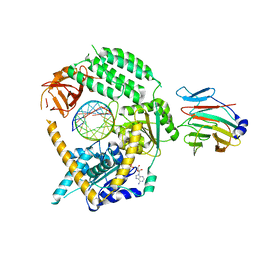 | | Cryo-EM structure of MDA5-dsRNA in complex with TRIM65 PSpry domain (Monomer) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Interferon-induced helicase C domain-containing protein 1, MAGNESIUM ION, ... | | Authors: | Kato, K, Ahmad, S, Hur, S. | | Deposit date: | 2020-07-29 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural analysis of RIG-I-like receptors reveals ancient rules of engagement between diverse RNA helicases and TRIM ubiquitin ligases.
Mol.Cell, 81, 2021
|
|
