4BLI
 
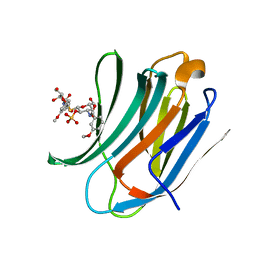 | | Galectin-3c in complex with Bisamido-thiogalactoside derivate 1 | | Descriptor: | (3-Deoxy-3-(3-methoxy-benzamido)-b-D-galactopyranosyl)-(3-deoxy-3-(3-methoxy-benzamido)-2-O-sulfo-b-D-galactopyranosyl)-sulfide, GALECTIN-3 | | Authors: | Noresson, A.L, Oberg, C.T, Engstrom, O, Hakansson, M, Logan, D.T, Leffler, H, Nilsson, U.J. | | Deposit date: | 2013-05-03 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Controlling Protein Conformation Through Electronic Fine-Tuning of Arginine-Arene Interactions: Synthetic, Structural, and Biological Studies
To be Published
|
|
6HYG
 
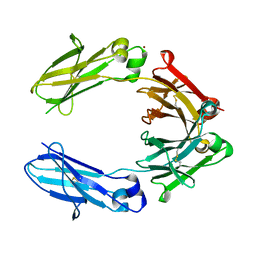 | | Heteromeric tandem IgG4/IgG1 Fc | | Descriptor: | IgHG1 and IgHG4 hybrid, ZINC ION | | Authors: | Casaletto, J.B, Geddie, M.L, Abu-Yousif, A.O, Masson, K, Fulgham, A, Boudot, A, Maiwald, T, Kearns, J.D, Kohli, N, Su, S, Razlog, M, Raue, A, Kalra, A, Hakansson, M, Logan, D.T, Welin, M, Chattopadhyay, S, Harms, B.D, Nielsen, U.B, Schoeberl, B, Lugovskoy, A.A, MacBeath, G. | | Deposit date: | 2018-10-21 | | Release date: | 2019-03-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | MM-131, a bispecific anti-Met/EpCAM mAb, inhibits HGF-dependent and HGF-independent Met signaling through concurrent binding to EpCAM.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
4BCZ
 
 | | Monomeric Human Cu,Zn Superoxide dismutase, loops IV and VII deleted, apo form. | | Descriptor: | SUPEROXIDE DISMUTASE [CU-ZN] | | Authors: | Saraboji, K, Awad, W, Danielsson, J, Lang, L, Kurnik, M, Marklund, S.L, Oliveberg, M, Logan, D.T. | | Deposit date: | 2012-10-03 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Global Structural Motions from the Strain of a Single Hydrogen Bond.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6I69
 
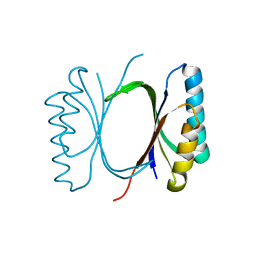 | |
6I6O
 
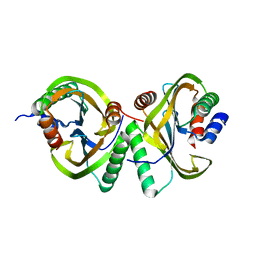 | | Circular permutant of ribosomal protein S6, swap helix 2, L75A mutant | | Descriptor: | 30S ribosomal protein S6,30S ribosomal protein S6 | | Authors: | Wang, H, Logan, D.T, Oliveberg, M. | | Deposit date: | 2018-11-15 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Exposing the distinctive modular behavior of beta-strands and alpha-helices in folded proteins.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6I6Y
 
 | |
6I77
 
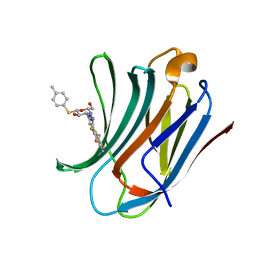 | | Galectin-3C in complex with substituted polyfluoroaryl monothiogalactoside derivative-4 | | Descriptor: | (2~{R},3~{R},4~{S},5~{R},6~{S})-4-[4-[4-azanyl-2,3,5,6-tetrakis(fluoranyl)phenyl]-1,2,3-triazol-1-yl]-2-(hydroxymethyl)-6-(4-methylphenyl)sulfanyl-oxane-3,5-diol, Galectin-3 | | Authors: | Kumar, R, Peterson, K, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2018-11-15 | | Release date: | 2019-01-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.219 Å) | | Cite: | Substituted polyfluoroaryl interactions with an arginine side chain in galectin-3 are governed by steric-, desolvation and electronic conjugation effects.
Org. Biomol. Chem., 17, 2019
|
|
6IC5
 
 | | Human cathepsin-C in complex with dipeptidyl cyclopropyl nitrile inhibitor 2 | | Descriptor: | (2~{S})-2-azanyl-~{N}-[(1~{R},2~{R})-1-(iminomethyl)-2-[4-[4-(4-methylpiperazin-1-yl)sulfonylphenyl]phenyl]cyclopropyl]-3-thiophen-2-yl-propanamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Hakansson, M, Logan, D.T, Korkmaz, B, Lesner, A, Wysocka, M, Gieldon, A, Gauthier, F, Jenne, D, Lauritzen, C, Pedersen, J. | | Deposit date: | 2018-12-02 | | Release date: | 2019-04-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design and in vivo anti-arthritic activity evaluation of a potent dipeptidyl cyclopropyl nitrile inhibitor of cathepsin C.
Biochem. Pharmacol., 164, 2019
|
|
6I07
 
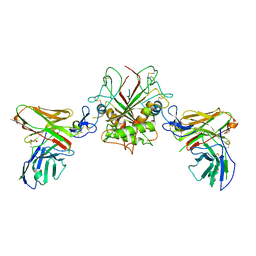 | | Crystal structure of EpCAM in complex with scFv | | Descriptor: | Epithelial cell adhesion molecule, GLYCEROL, Single chain Fv | | Authors: | Casaletto, J.B, Geddie, M.L, Abu-Yousif, A.O, Masson, K, Fulgham, A, Boudot, A, Maiwald, T, Kearns, J.D, Kohli, N, Su, S, Razlog, M, Raue, A, Kalra, A, Hakansson, M, Logan, D.T, Welin, M, Chattopadhyay, S, Harms, B.D, Nielsen, U.B, Schoeberl, B, Lugovskoy, A.A, MacBeath, G. | | Deposit date: | 2018-10-25 | | Release date: | 2019-03-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | MM-131, a bispecific anti-Met/EpCAM mAb, inhibits HGF-dependent and HGF-independent Met signaling through concurrent binding to EpCAM.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6I6I
 
 | | Circular permutant of ribosomal protein S6, adding 6aa to C terminal of P68-69, L75A mutant | | Descriptor: | 30S ribosomal protein S6,30S ribosomal protein S6, SULFATE ION | | Authors: | Wang, H, Logan, D.T, Oliveberg, M. | | Deposit date: | 2018-11-15 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Exposing the distinctive modular behavior of beta-strands and alpha-helices in folded proteins.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4AD7
 
 | | Crystal structure of full-length N-glycosylated human glypican-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYPICAN-1 | | Authors: | Svensson, G, Awad, W, Mani, K, Logan, D.T. | | Deposit date: | 2011-12-22 | | Release date: | 2012-03-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.945 Å) | | Cite: | Crystal Structure of N-Glycosylated Human Glypican-1 Core Protein: Structure of Two Loops Evolutionarily Conserved in Vertebrate Glypican-1.
J.Biol.Chem., 287, 2012
|
|
6I75
 
 | | Galectin-3C in complex with substituted polyfluoroaryl monothiogalactoside derivative 2 | | Descriptor: | (2~{R},3~{R},4~{S},5~{R},6~{S})-2-(hydroxymethyl)-6-(4-methylphenyl)sulfanyl-4-[4-[2,3,5,6-tetrakis(fluoranyl)-4-oxidanyl-phenyl]-1,2,3-triazol-1-yl]oxane-3,5-diol, Galectin-3 | | Authors: | Kumar, R, Peterson, K, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2018-11-15 | | Release date: | 2019-01-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.171 Å) | | Cite: | Substituted polyfluoroaryl interactions with an arginine side chain in galectin-3 are governed by steric-, desolvation and electronic conjugation effects.
Org. Biomol. Chem., 17, 2019
|
|
6I6S
 
 | | Circular permutant of ribosomal protein S6, adding 9aa to C terminal of P68-69, L75A mutant | | Descriptor: | 30S ribosomal protein S6,30S ribosomal protein S6,30S ribosomal protein S6,30S ribosomal protein S6,30S ribosomal protein S6,30S ribosomal protein S6,30S ribosomal protein S6,30S ribosomal protein S6, POTASSIUM ION, SODIUM ION | | Authors: | Wang, H, Logan, D.T, Oliveberg, M. | | Deposit date: | 2018-11-15 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Exposing the distinctive modular behavior of beta-strands and alpha-helices in folded proteins.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4BM8
 
 | | Galectin-3c in complex with Bisamido-thiogalactoside derivate 3 | | Descriptor: | (3-Deoxy-3-(2,3,5,6-tetra-fluoro-4-methoxy-benzamido)-b-D-galactopyranosyl)-(3-deoxy-3-(2,3,5,6-tetra-fluoro-4-methoxy-benzamido)-2-O-sulfo-b-D-galactopyranosyl)-sulfide, GALECTIN-3 | | Authors: | Noresson, A.L, Oberg, C.T, Engstrom, O, Hakansson, M, Logan, D.T, Leffler, H, Nilsson, U.J. | | Deposit date: | 2013-05-07 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | Controlling Protein Conformation Through Electronic Fine-Tuning of Arginine-Arene Interactions: Synthetic, Structural, and Biological Studies
To be Published
|
|
3ZSL
 
 | | Crystal structure of Apo Human Galectin-3 CRD at 1.08 angstrom resolution, at cryogenic temperature | | Descriptor: | GALECTIN-3 | | Authors: | Saraboji, K, Hakansson, M, Diehl, C, Nilsson, U.J, Leffler, H, Akke, M, Logan, D.T. | | Deposit date: | 2011-06-28 | | Release date: | 2011-12-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | The Carbohydrate-Binding Site in Galectin-3 is Pre-Organized to Recognize a Sugar-Like Framework of Oxygens: Ultra-High Resolution Structures and Water Dynamics.
Biochemistry, 51, 2012
|
|
4ACR
 
 | | Crystal structure of N-glycosylated, C-terminally truncated human glypican-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYPICAN-1 | | Authors: | Svensson, G, Awad, W, Mani, K, Logan, D.T. | | Deposit date: | 2011-12-17 | | Release date: | 2012-02-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal Structure of N-Glycosylated Human Glypican-1 Core Protein: Structure of Two Loops Evolutionarily Conserved in Vertebrate Glypican-1.
J.Biol.Chem., 287, 2012
|
|
4BCY
 
 | | Monomeric Human Cu,Zn Superoxide dismutase, mutation H43F | | Descriptor: | CADMIUM ION, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Awad, W, Saraboji, K, Danielsson, J, Lang, L, Kurnik, M, Marklund, S.L, Oliveberg, M, Logan, D.T. | | Deposit date: | 2012-10-03 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.272 Å) | | Cite: | Global Structural Motions from the Strain of a Single Hydrogen Bond.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4BLJ
 
 | | Galectin-3c in complex with Bisamido-thiogalactoside derivate 2 | | Descriptor: | Bis-(3-deoxy-3-(3-methoxy-benzamido)-b-D-galactopyranosyl)-sulfide, GALECTIN-3 | | Authors: | Noresson, A.L, Oberg, C.T, Engstrom, O, Hakansson, M, Logan, D.T, Leffler, H, Nilsson, U.J. | | Deposit date: | 2013-05-03 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Controlling Protein Conformation Through Electronic Fine-Tuning of Arginine-Arene Interactions: Synthetic, Structural, and Biological Studies
To be Published
|
|
6I6U
 
 | | Circular permutant of ribosomal protein S6, adding 9aa to N terminal of P81-82, L75A mutant | | Descriptor: | 30S ribosomal protein S6,30S ribosomal protein S6 | | Authors: | Wang, H, Logan, D.T, Oliveberg, M. | | Deposit date: | 2018-11-15 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Exposing the distinctive modular behavior of beta-strands and alpha-helices in folded proteins.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
3ZSJ
 
 | | Crystal structure of Human Galectin-3 CRD in complex with Lactose at 0.86 angstrom resolution | | Descriptor: | GALECTIN-3, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Saraboji, K, Hakansson, M, Diehl, C, Nilsson, U.J, Leffler, H, Akke, M, Logan, D.T. | | Deposit date: | 2011-06-28 | | Release date: | 2011-12-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (0.86 Å) | | Cite: | The Carbohydrate-Binding Site in Galectin-3 is Pre-Organized to Recognize a Sugar-Like Framework of Oxygens: Ultra-High Resolution Structures and Water Dynamics.
Biochemistry, 51, 2012
|
|
3ZSK
 
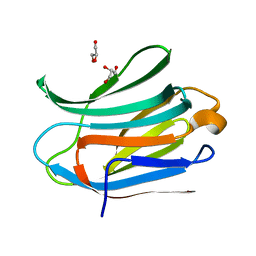 | | Crystal structure of Human Galectin-3 CRD with glycerol bound at 0.90 angstrom resolution | | Descriptor: | GALECTIN-3, GLYCEROL | | Authors: | Saraboji, K, Hakansson, M, Diehl, C, Nilsson, U.J, Leffler, H, Akke, M, Logan, D.T. | | Deposit date: | 2011-06-28 | | Release date: | 2011-12-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | The Carbohydrate-Binding Site in Galectin-3 is Pre-Organized to Recognize a Sugar-Like Framework of Oxygens: Ultra-High Resolution Structures and Water Dynamics.
Biochemistry, 51, 2012
|
|
4COJ
 
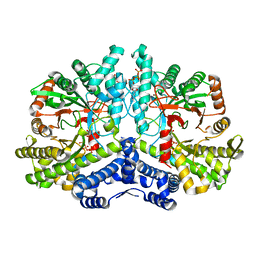 | | Crystal structure of the anaerobic ribonucleotide reductase from Thermotoga maritima in complex with dATP and CTP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, ANAEROBIC RIBONUCLEOSIDE-TRIPHOSPHATE REDUCTASE, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Aurelius, O, Johansson, R, Bagenholm, V, Beck, T, Balhuizen, A, Lundin, D, Sjoberg, B.M, Mulliez, E, Logan, D.T. | | Deposit date: | 2014-01-28 | | Release date: | 2015-05-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | The Crystal Structure of Thermotoga Maritima Class III Ribonucleotide Reductase Lacks a Radical Cysteine Pre-Positioned in the Active Site.
Plos One, 10, 2015
|
|
4COI
 
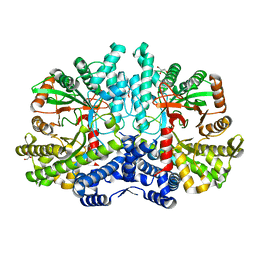 | | Crystal structure of the anaerobic ribonucleotide reductase from Thermotoga maritima with glycerol in the active site | | Descriptor: | ANAEROBIC RIBONUCLEOSIDE-TRIPHOSPHATE REDUCTASE, GLYCEROL, ZINC ION | | Authors: | Aurelius, O, Johansson, R, Bagenholm, V, Beck, T, Balhuizen, A, Lundin, D, Sjoberg, B.M, Mulliez, E, Logan, D.T. | | Deposit date: | 2014-01-28 | | Release date: | 2015-01-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | The Crystal Structure of Thermotoga Maritima Class III Ribonucleotide Reductase Lacks a Radical Cysteine Pre-Positioned in the Active Site.
Plos One, 10, 2015
|
|
6QGE
 
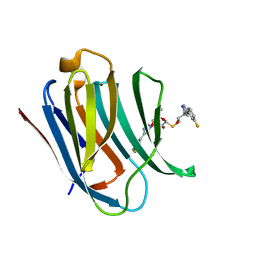 | | Galectin-3C in complex with a pair of enantiomeric ligands: S enantiomer | | Descriptor: | (2~{S},3~{R},4~{S},5~{R},6~{R})-4-[4-(3-fluorophenyl)-1,2,3-triazol-1-yl]-2-[(2~{S})-3-[4-(3-fluorophenyl)-1,2,3-triazol-1-yl]-2-oxidanyl-propyl]sulfanyl-6-(hydroxymethyl)oxane-3,5-diol, Galectin-3 | | Authors: | Manzoni, F, Verteramo, M.L, Oksanen, E, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2019-01-11 | | Release date: | 2019-01-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Interplay between Conformational Entropy and Solvation Entropy in Protein-Ligand Binding.
J. Am. Chem. Soc., 141, 2019
|
|
6QGF
 
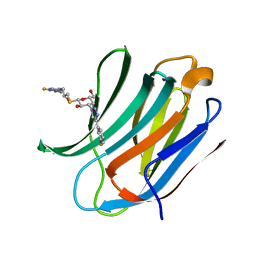 | | Galectin-3C in complex with a pair of enantiomeric ligands: R enantiomer | | Descriptor: | (2~{S},3~{R},4~{S},5~{R},6~{R})-4-[4-(3-fluorophenyl)-1,2,3-triazol-1-yl]-2-[(2~{R})-3-[4-(3-fluorophenyl)-1,2,3-triazol-1-yl]-2-oxidanyl-propyl]sulfanyl-6-(hydroxymethyl)oxane-3,5-diol, Galectin-3 | | Authors: | Manzoni, F, Verteramo, M.L, Oksanen, E, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2019-01-11 | | Release date: | 2019-01-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Interplay between Conformational Entropy and Solvation Entropy in Protein-Ligand Binding.
J. Am. Chem. Soc., 141, 2019
|
|
