3WSO
 
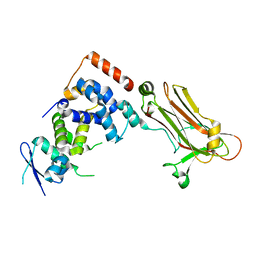 | | Crystal structure of the Skp1-FBG3 complex | | Descriptor: | F-box only protein 44, S-phase kinase-associated protein 1 | | Authors: | Kumanomidou, T, Nishio, K, Takagi, K, Nakagawa, T, Suzuki, A, Yamane, T, Tokunaga, F, Iwai, K, Murakami, A, Yoshida, Y, Tanaka, K, Mizushima, T. | | Deposit date: | 2014-03-18 | | Release date: | 2015-03-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Structural Differences between a Glycoprotein Specific F-Box Protein Fbs1 and Its Homologous Protein FBG3
Plos One, 10, 2015
|
|
5GWT
 
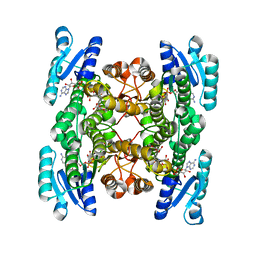 | | 4-hydroxyisoleucine dehydrogenase mutant complexed with NADH and succinate | | Descriptor: | 4-hydroxyisolecuine dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SUCCINIC ACID | | Authors: | Shi, X, Miyakawa, T, Nakamura, A, Tanokura, M. | | Deposit date: | 2016-09-13 | | Release date: | 2017-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Engineering a short-chain dehydrogenase/reductase for the stereoselective production of (2S,3R,4S)-4-hydroxyisoleucine with three asymmetric centers.
Sci Rep, 7, 2017
|
|
5GWR
 
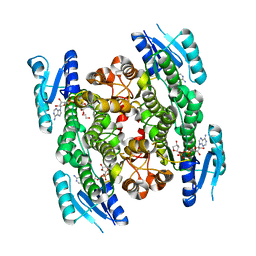 | | 4-hydroxyisoleucine dehydrogenase complexed with NADH | | Descriptor: | 1,2-ETHANEDIOL, 4-hydroxyisolecuine dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Shi, X, Miyakawa, T, Nakamura, A, Tanokura, M. | | Deposit date: | 2016-09-13 | | Release date: | 2017-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Engineering a short-chain dehydrogenase/reductase for the stereoselective production of (2S,3R,4S)-4-hydroxyisoleucine with three asymmetric centers
Sci Rep, 7, 2017
|
|
5GWS
 
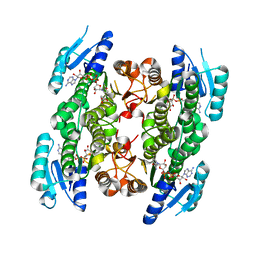 | | 4-hydroxyisoleucine dehydrogenase complexed with NADH and succinate | | Descriptor: | 4-hydroxyisolecuine dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SUCCINIC ACID | | Authors: | Shi, X, Miyakawa, T, Nakamura, A, Tanokura, M. | | Deposit date: | 2016-09-13 | | Release date: | 2017-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Engineering a short-chain dehydrogenase/reductase for the stereoselective production of (2S,3R,4S)-4-hydroxyisoleucine with three asymmetric centers.
Sci Rep, 7, 2017
|
|
3HR0
 
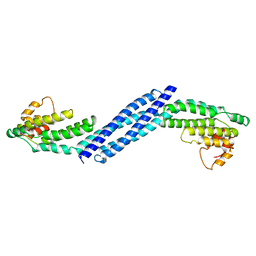 | | Crystal structure of Homo sapiens Conserved Oligomeric Golgi subunit 4 | | Descriptor: | CoG4 | | Authors: | Richardson, B.C, Ungar, D, Nakamura, A, Jeffrey, P.D, Hughson, F.M. | | Deposit date: | 2009-06-08 | | Release date: | 2009-07-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for a human glycosylation disorder caused by mutation of the COG4 gene.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
5H3F
 
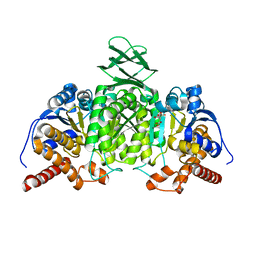 | | Crystal structure of mouse isocitrate dehydrogenases 2 complexed with isocitrate | | Descriptor: | ISOCITRIC ACID, Isocitrate dehydrogenase [NADP], mitochondrial, ... | | Authors: | Xu, Y, Liu, L, Miyakawa, T, Nakamura, A, Tanokura, M. | | Deposit date: | 2016-10-23 | | Release date: | 2017-08-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Studies on the regulatory mechanism of isocitrate dehydrogenase 2 using acetylation mimics
Sci Rep, 7, 2017
|
|
5H3E
 
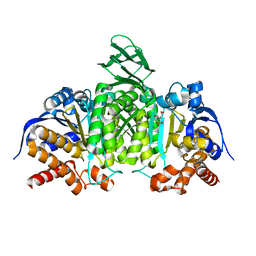 | | Crystal structure of mouse isocitrate dehydrogenases 2 K256Q mutant complexed with isocitrate | | Descriptor: | ISOCITRIC ACID, Isocitrate dehydrogenase [NADP], mitochondrial, ... | | Authors: | Xu, Y, Liu, L, Miyakawa, T, Nakamura, A, Tanokura, M. | | Deposit date: | 2016-10-23 | | Release date: | 2017-08-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Studies on the regulatory mechanism of isocitrate dehydrogenase 2 using acetylation mimics
Sci Rep, 7, 2017
|
|
7Y8V
 
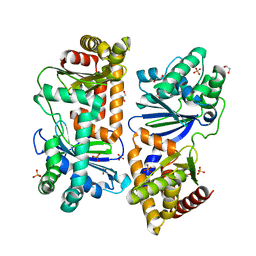 | | Crystal structure of AlbEF homolog mutant (AlbF-H54A/H58A) from Quasibacillus thermotolerans | | Descriptor: | 1,2-ETHANEDIOL, AlbE homolog, AlbF homolog H54A/H58A mutant, ... | | Authors: | Ishida, K, Nakamura, A, Kojima, S. | | Deposit date: | 2022-06-24 | | Release date: | 2022-10-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the AlbEF complex involved in subtilosin A biosynthesis.
Structure, 30, 2022
|
|
6RWY
 
 | |
7Y8U
 
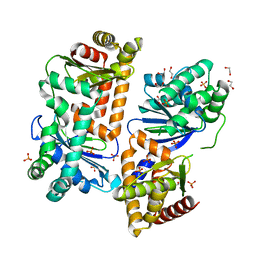 | | Crystal structure of AlbEF homolog from Quasibacillus thermotolerans | | Descriptor: | 1,2-ETHANEDIOL, AlbE homolog, AlbF homolog, ... | | Authors: | Ishida, K, Nakamura, A, Kojima, S. | | Deposit date: | 2022-06-24 | | Release date: | 2022-10-12 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the AlbEF complex involved in subtilosin A biosynthesis.
Structure, 30, 2022
|
|
7Y8X
 
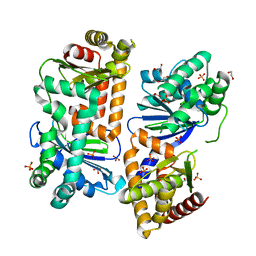 | | Crystal structure of AlbEF homolog from Quasibacillus thermotolerans in complex with Ni(II) | | Descriptor: | 1,2-ETHANEDIOL, AlbE homolog, AlbF homolog, ... | | Authors: | Ishida, K, Nakamura, A, Kojima, S. | | Deposit date: | 2022-06-24 | | Release date: | 2022-10-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of the AlbEF complex involved in subtilosin A biosynthesis.
Structure, 30, 2022
|
|
3IYZ
 
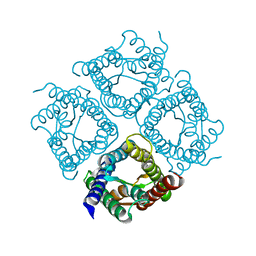 | | Structure of Aquaporin-4 S180D mutant at 10.0 A resolution from electron micrograph | | Descriptor: | Aquaporin-4 | | Authors: | Mitsuma, T, Tani, K, Hiroaki, Y, Kamegawa, A, Suzuki, H, Hibino, H, Kurachi, Y, Fujiyoshi, Y. | | Deposit date: | 2010-07-24 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | ELECTRON CRYSTALLOGRAPHY (10 Å) | | Cite: | Influence of the cytoplasmic domains of aquaporin-4 on water conduction and array formation.
J.Mol.Biol., 402, 2010
|
|
2KTT
 
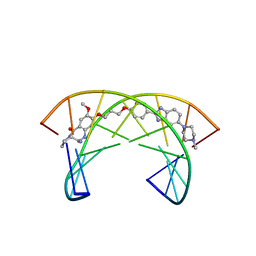 | | Solution Structure of a Covalently Bound Pyrrolo[2,1-c][1,4]benzodiazepine-Benzimidazole Hybrid to a 10mer DNA Duplex | | Descriptor: | (11aS)-7-methoxy-8-(3-{4-[6-(4-methylpiperazin-1-yl)-1H-benzimidazol-2-yl]phenoxy}propoxy)-1,2,3,10,11,11a-hexahydro-5H-pyrrolo[2,1-c][1,4]benzodiazepin-5-one, 5'-D(*AP*AP*CP*AP*AP*TP*TP*GP*TP*T)-3' | | Authors: | Rettig, M, Weingarth, M, Langel, W, Kamal, A, Kumar, P.P, Weisz, K. | | Deposit date: | 2010-02-08 | | Release date: | 2010-03-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a covalently bound pyrrolo[2,1-c][1,4]benzodiazepine-benzimidazole hybrid to a 10mer DNA duplex.
Biochemistry, 48, 2009
|
|
9IHS
 
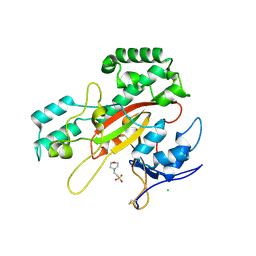 | | Microbial transglutaminase mutant - D3C/G283C | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Suzuki, M, Date, M, Kashiwagi, T, Takahashi, K, Nakamura, A, Tanokura, M, Suzuki, E, Yokoyama, K. | | Deposit date: | 2024-06-18 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Random mutagenesis and disulfide bond formation improved thermostability in microbial transglutaminase.
Appl.Microbiol.Biotechnol., 108, 2024
|
|
7WMT
 
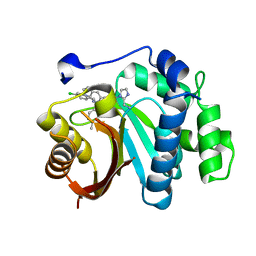 | | Crystal structure of small molecule 13 bound to human Nicotinamide N-methyltransferase | | Descriptor: | Nicotinamide N-methyltransferase, [(2~{R},4~{S})-4-[2-(aminomethyl)imidazol-1-yl]-2-[1-[(4-chlorophenyl)methyl]-5-methyl-indol-2-yl]pyrrolidin-1-yl]-(1~{H}-pyrrolo[2,3-b]pyridin-5-yl)methanone | | Authors: | Yoshida, S, Uehara, S, Kondo, N, Takahashi, Y, Yamamoto, S, Kameda, A, Kawagoe, S, Inoue, N, Yamada, M, Yoshimura, N, Tachibana, Y. | | Deposit date: | 2022-01-17 | | Release date: | 2022-08-31 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Peptide-to-Small Molecule: A Pharmacophore-Guided Small Molecule Lead Generation Strategy from High-Affinity Macrocyclic Peptides.
J.Med.Chem., 65, 2022
|
|
7WMC
 
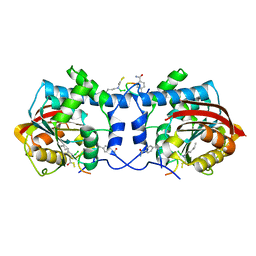 | | Crystal structure of macrocyclic peptide 1 bound to human Nicotinamide N-methyltransferase | | Descriptor: | Nicotinamide N-methyltransferase, Peptide1 | | Authors: | Yoshida, S, Uehara, S, Kondo, N, Takahashi, Y, Yamamoto, S, Kameda, A, Kawagoe, S, Inoue, N, Yamada, M, Yoshimura, N, Tachibana, Y. | | Deposit date: | 2022-01-14 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Peptide-to-Small Molecule: A Pharmacophore-Guided Small Molecule Lead Generation Strategy from High-Affinity Macrocyclic Peptides.
J.Med.Chem., 65, 2022
|
|
1VGG
 
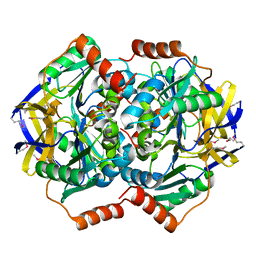 | | Crystal Structure of the Conserved Hypothetical Protein TTHA1091 from Thermus Thermophilus HB8 | | Descriptor: | Conserved Hypothetical Protein TT1634 (TTHA1091) | | Authors: | Satoh, S, Yao, M, Kousumi, Y, Ebihara, A, Matsumoto, K, Okamoto, A, Tanaka, I, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-04-26 | | Release date: | 2004-10-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of the Conserved Hypothetical Protein TT1634 from Thermus Thermophilus HB8
To be Published
|
|
1VDH
 
 | | Structure-based functional identification of a novel heme-binding protein from thermus thermophilus HB8 | | Descriptor: | muconolactone isomerase-like protein | | Authors: | Ebihara, A, Okamoto, A, Kousumi, Y, Yamamoto, H, Masui, R, Ueyama, N, Shibata, T, Inoue, Y, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-03-22 | | Release date: | 2004-09-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based functional identification of a novel heme-binding protein from Thermus thermophilus HB8.
J.STRUCT.FUNCT.GENOM., 6, 2005
|
|
5WUT
 
 | |
9J7I
 
 | | Cryo-EM Structure of calcium sensing receptor in complex gamma-glutamyl-valyl-glycine as a kokumi substance | | Descriptor: | Extracellular calcium-sensing receptor, gamma-glutamyl-valyl-glycine | | Authors: | Yamaguchi, H, Kitajima, S, Suzuki, H, Suzuki, S, Nishikawa, K, Maruyama, Y, Kamegawa, A, Kazutoshi, T, Tagami, U, Kuroda, M, Fujiyoshi, Y, Sugiki, M. | | Deposit date: | 2024-08-19 | | Release date: | 2025-02-19 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Cryo-EM structure of the calcium-sensing receptor complexed with the kokumi substance gamma-glutamyl-valyl-glycine.
Sci Rep, 15, 2025
|
|
5XCZ
 
 | | Structure of the cellobiohydrolase Cel6A from Phanerochaete chrysosporium in complex with cellobiose at 2.1 angstrom | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glucanase, beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Tachioka, M, Nakamura, A, Ishida, T, Igarashi, K, Samejima, M. | | Deposit date: | 2017-03-24 | | Release date: | 2017-07-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a family 6 cellobiohydrolase from the basidiomycete Phanerochaete chrysosporium
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
2KY7
 
 | | NMR Structural Studies on the Covalent DNA Binding of a Pyrrolobenzodiazepine-Naphthalimide Conjugate | | Descriptor: | 2-{2-[4-(3-{[(11aS)-7-methoxy-5-oxo-2,3,5,10,11,11a-hexahydro-1H-pyrrolo[2,1-c][1,4]benzodiazepin-8-yl]oxy}propyl)piperazin-1-yl]ethyl}-1H-benzo[de]isoquinoline-1,3(2H)-dione, 5'-D(*AP*AP*CP*AP*AP*TP*TP*GP*TP*T)-3' | | Authors: | Rettig, M, Langel, W, Kamal, A, Weisz, K. | | Deposit date: | 2010-05-17 | | Release date: | 2010-06-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR structural studies on the covalent DNA binding of a pyrrolobenzodiazepine-naphthalimide conjugate
Org.Biomol.Chem., 8, 2010
|
|
2DB3
 
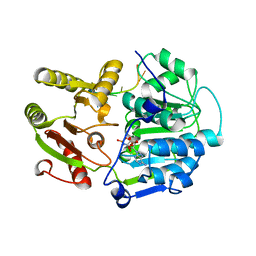 | | Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa | | Descriptor: | 5'-R(*UP*UP*UP*UP*UP*UP*UP*UP*UP*U)-3', ATP-dependent RNA helicase vasa, MAGNESIUM ION, ... | | Authors: | Sengoku, T, Nureki, O, Nakamura, A, Kobayashi, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-14 | | Release date: | 2006-05-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa.
Cell(Cambridge,Mass.), 125, 2006
|
|
4GA6
 
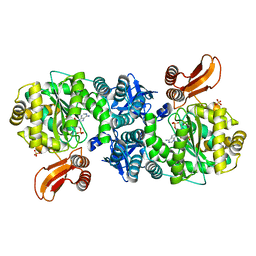 | | Crystal structure of AMP phosphorylase C-terminal deletion mutant in complex with substrates | | Descriptor: | ADENOSINE MONOPHOSPHATE, Putative thymidine phosphorylase, SULFATE ION | | Authors: | Nishitani, Y, Aono, R, Nakamura, A, Sato, T, Atomi, H, Imanaka, T, Miki, K. | | Deposit date: | 2012-07-25 | | Release date: | 2013-05-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structure analysis of archaeal AMP phosphorylase reveals two unique modes of dimerization
J.Mol.Biol., 425, 2013
|
|
5ZD4
 
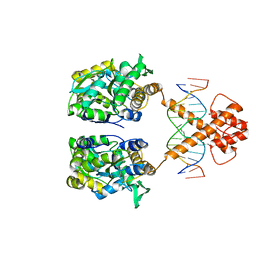 | | Crystal structure of MBP-fused BIL1/BZR1 in complex with double-stranded DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*TP*TP*CP*AP*CP*AP*CP*GP*TP*GP*TP*GP*AP*AP*A)-3'), Maltose-binding periplasmic protein,Protein BRASSINAZOLE-RESISTANT 1, ... | | Authors: | Nosaki, S, Miyakawa, T, Xu, Y, Nakamura, A, Hirabayashi, K, Tanokura, M. | | Deposit date: | 2018-02-22 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structural basis for brassinosteroid response by BIL1/BZR1.
Nat Plants, 4, 2018
|
|
