3IIG
 
 | |
3ILB
 
 | |
7E43
 
 | |
3IHD
 
 | |
3HWW
 
 | |
3IHF
 
 | |
3IIH
 
 | |
3IHE
 
 | |
3ILC
 
 | |
3IHC
 
 | |
3GNT
 
 | |
7CPZ
 
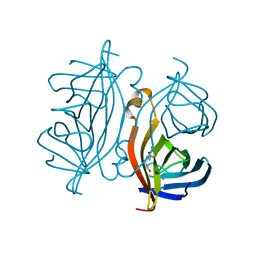 | | Crystal structure of Streptoavidin-C1 from Streptomyces cinamonensis | | Descriptor: | BIOTIN, Mature Streptoavidin-C1 | | Authors: | Jeon, B.J, Kim, S, Lee, J.-H, Kim, M.S, Hwang, K.Y. | | Deposit date: | 2020-08-08 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insights into the structure of mature streptavidin C1 from Streptomyces cinnamonensis reveal the self-binding of the extension C-terminal peptide to biotin-binding sites.
Iucrj, 8, 2021
|
|
7CQ0
 
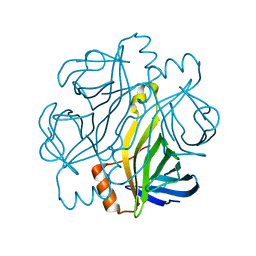 | | Crystal structure of Streptoavidin-C1 from Streptomyces cinamonensis | | Descriptor: | Mature Streptoavidin-C1 | | Authors: | Jeon, B.J, Kim, S, Lee, J.-H, Kim, M.S, Hwang, K.Y. | | Deposit date: | 2020-08-08 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Insights into the structure of mature streptavidin C1 from Streptomyces cinnamonensis reveal the self-binding of the extension C-terminal peptide to biotin-binding sites.
Iucrj, 8, 2021
|
|
3HWX
 
 | |
5X68
 
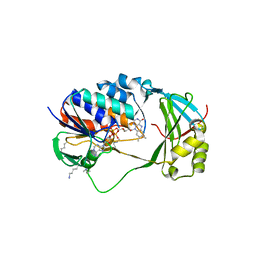 | | Crystal Structure of Human KMO | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Kynurenine 3-monooxygenase | | Authors: | Kim, H.T, Hwang, K.Y. | | Deposit date: | 2017-02-21 | | Release date: | 2018-02-21 | | Last modified: | 2018-05-02 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for Inhibitor-Induced Hydrogen Peroxide Production by Kynurenine 3-Monooxygenase
Cell Chem Biol, 25, 2018
|
|
5X6P
 
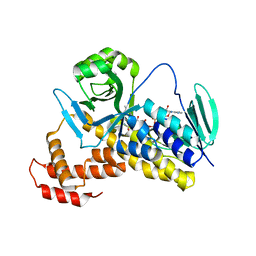 | | Crystal structure of Pseudomonas fluorescens KMO | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Kynurenine 3-monooxygenase | | Authors: | Kim, H.T, Hwang, K.Y. | | Deposit date: | 2017-02-22 | | Release date: | 2018-02-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural Basis for Inhibitor-Induced Hydrogen Peroxide Production by Kynurenine 3-Monooxygenase
Cell Chem Biol, 25, 2018
|
|
5XOX
 
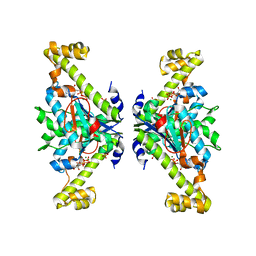 | | Crystal structure of tRNA(His) guanylyltranserase from Saccharomyces cerevisiae | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Lee, K, Lee, E.H, Son, J, Hwang, K.Y. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of tRNA(His) guanylyltransferase from Saccharomyces cerevisiae
Biochem. Biophys. Res. Commun., 490, 2017
|
|
5ZGA
 
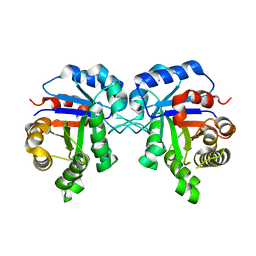 | | Crystal Structure of Triosephosphate isomerase SAD deletion and N115A mutant from Opisthorchis viverrini | | Descriptor: | Triosephosphate isomerase | | Authors: | Son, J, Kim, S, Kim, S.E, Lee, H, Lee, M.R, Hwang, K.Y. | | Deposit date: | 2018-03-08 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.793 Å) | | Cite: | Structural Analysis of an Epitope Candidate of Triosephosphate Isomerase in Opisthorchis viverrini.
Sci Rep, 8, 2018
|
|
5ZG5
 
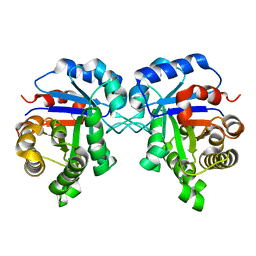 | | Crystal Structure of Triosephosphate isomerase SADsubAAA mutant from Opisthorchis viverrini | | Descriptor: | Triosephosphate isomerase | | Authors: | Son, J, Kim, S, Kim, S.E, Lee, H, Lee, M.R, Hwang, K.Y. | | Deposit date: | 2018-03-07 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.597 Å) | | Cite: | Structural Analysis of an Epitope Candidate of Triosephosphate Isomerase in Opisthorchis viverrini.
Sci Rep, 8, 2018
|
|
5ZG4
 
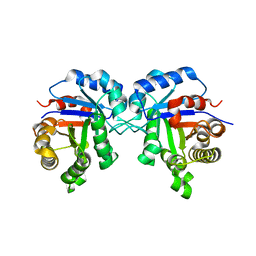 | | Crystal Structure of Triosephosphate isomerase SAD deletion mutant from Opisthorchis viverrini | | Descriptor: | Triosephosphate isomerase | | Authors: | Son, J, Kim, S, Kim, S.E, Lee, H, Lee, M.R, Hwang, K.Y. | | Deposit date: | 2018-03-07 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.746 Å) | | Cite: | Structural Analysis of an Epitope Candidate of Triosephosphate Isomerase in Opisthorchis viverrini.
Sci Rep, 8, 2018
|
|
5ZFX
 
 | | Crystal Structure of Triosephosphate isomerase from Opisthorchis viverrini | | Descriptor: | MAGNESIUM ION, Triosephosphate isomerase | | Authors: | Son, J, Kim, S, Kim, S.E, Lee, H, Lee, M.R, Hwang, K.Y. | | Deposit date: | 2018-03-07 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | Structural Analysis of an Epitope Candidate of Triosephosphate Isomerase in Opisthorchis viverrini.
Sci Rep, 8, 2018
|
|
6AA7
 
 | |
5X6Q
 
 | | Crystal structure of Pseudomonas fluorescens KMO in complex with Ro 61-8048 | | Descriptor: | 3,4-dimethoxy-N-[4-(3-nitrophenyl)-1,3-thiazol-2-yl]benzenesulfonamide, FLAVIN-ADENINE DINUCLEOTIDE, Kynurenine 3-monooxygenase | | Authors: | Kim, H.T, Hwang, K.Y. | | Deposit date: | 2017-02-23 | | Release date: | 2018-02-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.897 Å) | | Cite: | Structural Basis for Inhibitor-Induced Hydrogen Peroxide Production by Kynurenine 3-Monooxygenase
Cell Chem Biol, 25, 2018
|
|
5X6R
 
 | |
5IZV
 
 | | Crystal structure of the legionella pneumophila effector protein RavZ - F222 | | Descriptor: | Uncharacterized protein RavZ | | Authors: | Kwon, D.H, Kim, L, Kim, B.-W, Hong, S.B, Song, H.K. | | Deposit date: | 2016-03-26 | | Release date: | 2016-11-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.814 Å) | | Cite: | The 1:2 complex between RavZ and LC3 reveals a mechanism for deconjugation of LC3 on the phagophore membrane
Autophagy, 13, 2017
|
|
