8AFV
 
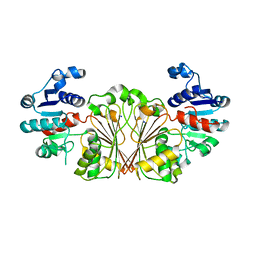 | | DaArgC3 - Engineered Formyl Phosphate Reductase with 3 substitutions (S178V, G182V, L233I) | | Descriptor: | N-acetyl-gamma-glutamyl-phosphate reductase, SODIUM ION | | Authors: | Pfister, P, Nattermann, M, Zarzycki, J, Erb, T.J. | | Deposit date: | 2022-07-18 | | Release date: | 2023-04-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Engineering a new-to-nature cascade for phosphate-dependent formate to formaldehyde conversion in vitro and in vivo.
Nat Commun, 14, 2023
|
|
8AFU
 
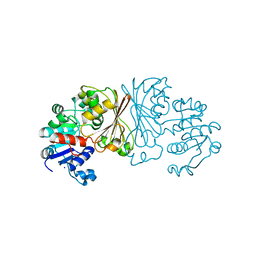 | | DaArgC - N-acetyl-gamma-glutamyl-phosphate Reductase of Denitrovibrio acetiphilus | | Descriptor: | N-acetyl-gamma-glutamyl-phosphate reductase, SODIUM ION | | Authors: | Pfister, P, Nattermann, M, Zarzycki, J, Erb, T.J. | | Deposit date: | 2022-07-18 | | Release date: | 2023-04-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Engineering a new-to-nature cascade for phosphate-dependent formate to formaldehyde conversion in vitro and in vivo.
Nat Commun, 14, 2023
|
|
8J45
 
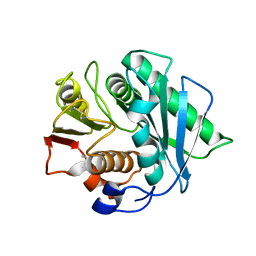 | | Crystal structure of a Pichia pastoris-expressed IsPETase variant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Poly(ethylene terephthalate) hydrolase | | Authors: | Li, X, He, H.L, Long, X, Niu, D, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-04-19 | | Release date: | 2024-01-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Complete decomposition of poly(ethylene terephthalate) by crude PET hydrolytic enzyme produced in Pichia pastoris
Chem Eng J, 2023
|
|
9ASI
 
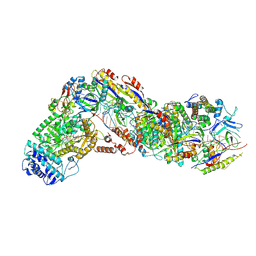 | | Cryo-EM structure of the active Lactococcus lactis Csm bound to target in pre-cleavage stage | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, CRISPR RNA, ... | | Authors: | Wang, B, Goswami, H.N, Li, H. | | Deposit date: | 2024-02-25 | | Release date: | 2024-08-14 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Molecular basis for cA6 synthesis by a type III-A CRISPR-Cas enzyme and its conversion to cA4 production.
Nucleic Acids Res., 52, 2024
|
|
9ASH
 
 | | Cryo-EM structure of the active Lactococcus lactis Csm bound to target in post-cleavage stage | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CRISPR RNA, CRISPR system Cms endoribonuclease Csm3, ... | | Authors: | Wang, B, Goswami, H.N, Li, H. | | Deposit date: | 2024-02-25 | | Release date: | 2024-08-14 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | Molecular basis for cA6 synthesis by a type III-A CRISPR-Cas enzyme and its conversion to cA4 production.
Nucleic Acids Res., 52, 2024
|
|
5GPK
 
 | | Crystal structure of Ccp1 mutant | | Descriptor: | Putative nucleosome assembly protein C36B7.08c | | Authors: | Yin, F, Gao, F, Chen, Y. | | Deposit date: | 2016-08-03 | | Release date: | 2016-11-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | Ccp1 Homodimer Mediates Chromatin Integrity by Antagonizing CENP-A Loading
Mol.Cell, 64, 2016
|
|
4M8M
 
 | |
5GPL
 
 | | Crystal structure of Ccp1 | | Descriptor: | Putative nucleosome assembly protein C36B7.08c | | Authors: | Yin, F, Gao, F, Chen, Y. | | Deposit date: | 2016-08-03 | | Release date: | 2016-11-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Ccp1 Homodimer Mediates Chromatin Integrity by Antagonizing CENP-A Loading
Mol.Cell, 64, 2016
|
|
2IIT
 
 | | Human dipeptidyl peptidase 4 in complex with a diazepan-2-one inhibitor | | Descriptor: | (3R)-4-[(3R)-3-AMINO-4-(2,4,5-TRIFLUOROPHENYL)BUTANOYL]-3-(2,2,2-TRIFLUOROETHYL)-1,4-DIAZEPAN-2-ONE, 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Scapin, G, Biftu, T, Weber, A.E. | | Deposit date: | 2006-09-28 | | Release date: | 2006-11-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | (3R)-4-[(3R)-3-Amino-4-(2,4,5-trifluorophenyl)butanoyl]-3-(2,2,2-trifluoroethyl)-1,4-diazepan-2-one, a selective dipeptidyl peptidase IV inhibitor for the treatment of type 2 diabetes
Bioorg.Med.Chem.Lett., 17, 2007
|
|
5Z2C
 
 | | Crystal structure of ALPK-1 N-terminal domain in complex with ADP-heptose | | Descriptor: | Alpha-protein kinase 1, [[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2S,3S,4S,5S,6R)-6-[(1S)-1,2-bis(oxidanyl)ethyl]-3,4,5-tris(oxidanyl)oxan-2-yl] hydrogen phosphate | | Authors: | Ding, J, She, Y, Shao, F. | | Deposit date: | 2018-01-02 | | Release date: | 2018-08-22 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.594 Å) | | Cite: | Alpha-kinase 1 is a cytosolic innate immune receptor for bacterial ADP-heptose.
Nature, 561, 2018
|
|
5WDH
 
 | | Motor domain of human kinesin family member C1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIFC1, MAGNESIUM ION, ... | | Authors: | Zhu, H, Tempel, W, He, H, Shen, Y, Wang, J, Brothers, G, Landry, R, Arrowsmith, C.H, Edwards, A.M, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-07-05 | | Release date: | 2017-08-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.248 Å) | | Cite: | Structural basis of small molecule ATPase inhibition of a human mitotic kinesin motor protein.
Sci Rep, 7, 2017
|
|
2HHA
 
 | | The structure of DPP4 in complex with an oxadiazole inhibitor | | Descriptor: | (2S,3S)-3-{3-[4-(METHYLSULFONYL)PHENYL]-1,2,4-OXADIAZOL-5-YL}-1-OXO-1-PYRROLIDIN-1-YLBUTAN-2-AMINE, 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Scapin, G. | | Deposit date: | 2006-06-28 | | Release date: | 2006-09-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Discovery of potent, selective, and orally bioavailable oxadiazole-based dipeptidyl peptidase IV inhibitors.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2IIV
 
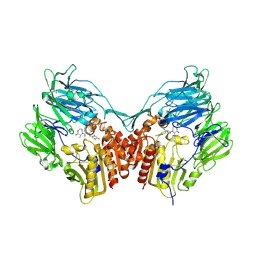 | | Human dipeptidyl peptidase 4 in complex with a diazepan-2-one inhibitor | | Descriptor: | (3R)-4-[(3R)-3-AMINO-4-(2,4,5-TRIFLUOROPHENYL)BUTANOYL]-3-METHYL-1,4-DIAZEPAN-2-ONE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Scapin, G, Weber, A.E, Biftu, T. | | Deposit date: | 2006-09-28 | | Release date: | 2006-11-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | (3R)-4-[(3R)-3-Amino-4-(2,4,5-trifluorophenyl)butanoyl]-3-(2,2,2-trifluoroethyl)-1,4-diazepan-2-one, a selective dipeptidyl peptidase IV inhibitor for the treatment of type 2 diabetes
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2OPH
 
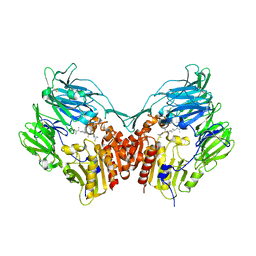 | | Human dipeptidyl peptidase IV in complex with an alpha amino acid inhibitor | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Scapin, G, Weber, A.E, Duffy, J.L. | | Deposit date: | 2007-01-29 | | Release date: | 2007-05-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 4-Aminophenylalanine and 4-aminocyclohexylalanine derivatives as potent, selective, and orally bioavailable inhibitors of dipeptidyl peptidase IV.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
8X2J
 
 | |
9HNF
 
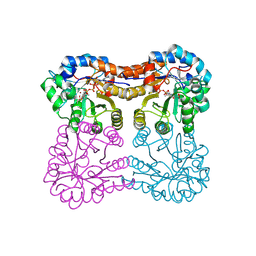 | | Beta-keto acid cleavage enzyme from Paracoccus denitrificans with bound acetoacetate and acetyl-CoA | | Descriptor: | 3-keto-5-aminohexanoate cleavage protein, ACETOACETIC ACID, ACETYL COENZYME *A, ... | | Authors: | Marchal, D.G, Zarzycki, J, Erb, T.J. | | Deposit date: | 2024-12-10 | | Release date: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design and implementation of aerobic and ambient CO 2 -reduction as an entry-point for enhanced carbon fixation.
Nat Commun, 16, 2025
|
|
2P8S
 
 | | Human dipeptidyl peptidase IV/CD26 in complex with a cyclohexalamine inhibitor | | Descriptor: | (1S,2R,5S)-5-[3-(TRIFLUOROMETHYL)-5,6-DIHYDRO[1,2,4]TRIAZOLO[4,3-A]PYRAZIN-7(8H)-YL]-2-(2,4,5-TRIFLUOROPHENYL)CYCLOHEXANAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Scapin, G, Biftu, T. | | Deposit date: | 2007-03-23 | | Release date: | 2007-05-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Rational design of a novel, potent, and orally bioavailable cyclohexylamine DPP-4 inhibitor by application of molecular modeling and X-ray crystallography of sitagliptin
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2QOE
 
 | | Human Dipeptidyl Peptidase IV in complex with a Triazolopiperazine-based beta amino acid Inhibitor | | Descriptor: | (2R)-4-[(8R)-8-METHYL-2-(TRIFLUOROMETHYL)-5,6-DIHYDRO[1,2,4]TRIAZOLO[1,5-A]PYRAZIN-7(8H)-YL]-4-OXO-1-(2,4,5-TRIFLUOROPHENYL)BUTAN-2-AMINE, 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Scapin, G. | | Deposit date: | 2007-07-20 | | Release date: | 2007-11-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design, synthesis, and biological evaluation of triazolopiperazine-based beta-amino amides as potent, orally active dipeptidyl peptidase IV (DPP-4) inhibitors.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
7V4Q
 
 | |
7EI1
 
 | | Structure of Pyrococcus furiosus Cas1Cas2 complex | | Descriptor: | CRISPR-associated endonuclease Cas1, CRISPR-associated endoribonuclease Cas2 | | Authors: | Yu, Y, Chen, Q. | | Deposit date: | 2021-03-30 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | A distinct structure of Cas1-Cas2 complex provides insights into the mechanism for the longer spacer acquisition in Pyrococcus furiosus.
Int.J.Biol.Macromol., 183, 2021
|
|
7V4R
 
 | | The crystal structure of KFDV NS3H bound with Pi | | Descriptor: | NICKEL (II) ION, PHOSPHATE ION, Serine protease NS3 | | Authors: | Zhang, C.Y, Jin, T.C. | | Deposit date: | 2021-08-14 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Kyasanur Forest disease virus NS3 helicase: Insights into structure, activity, and inhibitors.
Int.J.Biol.Macromol., 2023
|
|
3I2N
 
 | | Crystal Structure of WD40 repeats protein WDR92 | | Descriptor: | WD repeat-containing protein 92 | | Authors: | Amaya, M.F, Li, Z, He, H, Seitova, A, Ni, S, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Bountra, C, Bochkarev, A, Min, J, Ouyang, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-06-29 | | Release date: | 2009-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure and function of WD40 domain proteins.
Protein Cell, 2, 2011
|
|
3GFC
 
 | | Crystal Structure of Histone-binding protein RBBP4 | | Descriptor: | Histone-binding protein RBBP4 | | Authors: | Amaya, M.F, Dong, A, Li, Z, He, H, Ni, S, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Bountra, C, Bochkarev, A, Min, J, Ouyang, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-02-26 | | Release date: | 2009-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and function of WD40 domain proteins.
Protein Cell, 2, 2011
|
|
1GHD
 
 | | Crystal structure of the glutaryl-7-aminocephalosporanic acid acylase by mad phasing | | Descriptor: | GLUTARYL-7-AMINOCEPHALOSPORANIC ACID ACYLASE | | Authors: | Ding, Y, Jiang, W, Mao, X, He, H, Zhang, S, Tang, H, Bartlam, M, Ye, S, Jiang, F, Liu, Y, Zhao, G, Rao, Z. | | Deposit date: | 2000-12-07 | | Release date: | 2003-07-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Affinity alkylation of the Trp-B4 residue of the beta -subunit of the glutaryl 7-aminocephalosporanic acid acylase of Pseudomonas sp. 130.
J.Biol.Chem., 277, 2002
|
|
8GI5
 
 | |
