7JL4
 
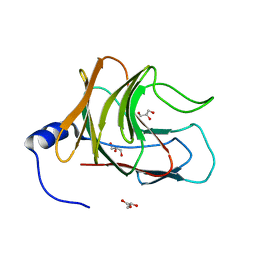 | | Crystal structure of TRIM65 PSpry domain | | Descriptor: | GLYCEROL, Tripartite motif-containing protein 65 | | Authors: | Kato, K, Ahmad, S, Hur, S. | | Deposit date: | 2020-07-29 | | Release date: | 2020-12-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural analysis of RIG-I-like receptors reveals ancient rules of engagement between diverse RNA helicases and TRIM ubiquitin ligases.
Mol.Cell, 81, 2021
|
|
7JL2
 
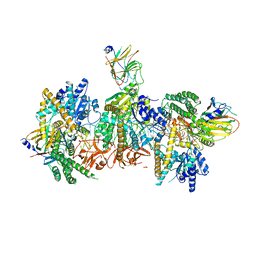 | | Cryo-EM structure of MDA5-dsRNA filament in complex with TRIM65 PSpry domain (Trimer) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Interferon-induced helicase C domain-containing protein 1, MAGNESIUM ION, ... | | Authors: | Kato, K, Ahmad, S, Hur, S. | | Deposit date: | 2020-07-29 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural analysis of RIG-I-like receptors reveals ancient rules of engagement between diverse RNA helicases and TRIM ubiquitin ligases.
Mol.Cell, 81, 2021
|
|
6JZB
 
 | |
4L0K
 
 | |
4LG8
 
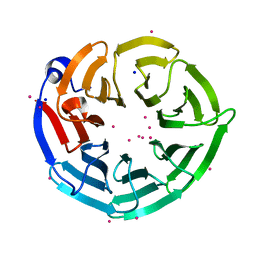 | | Crystal structure of PRPF19 WD40 repeats | | Descriptor: | Pre-mRNA-processing factor 19, SODIUM ION, UNKNOWN ATOM OR ION | | Authors: | Xu, C, Tempel, W, He, H, Dobrovetsky, E, Seitova, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-06-27 | | Release date: | 2013-08-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structure of the WD40 domain of human PRPF19.
Biochem. Biophys. Res. Commun., 493, 2017
|
|
8DSR
 
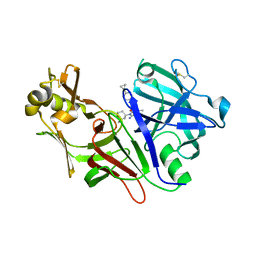 | | Structure of Plasmepsin X (PM10, PMX) from Plasmodium falciparum 3D7 in complex with UCB7362 | | Descriptor: | (2E,6S)-6-{2-chloro-3-[(2-cyclopropylpyrimidin-5-yl)amino]phenyl}-2-imino-6-methyl-3-[(2S,4S)-2-methyloxan-4-yl]-1,3-diazinan-4-one, Plasmepsin X | | Authors: | Abendroth, J, Lorimer, D.D. | | Deposit date: | 2022-07-22 | | Release date: | 2022-10-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Discovery and Characterization of Potent, Efficacious, Orally Available Antimalarial Plasmepsin X Inhibitors and Preclinical Safety Assessment of UCB7362 .
J.Med.Chem., 65, 2022
|
|
3WCA
 
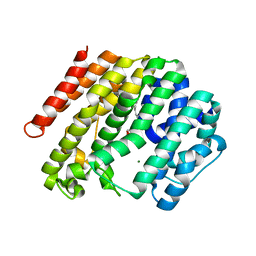 | | The complex structure of TcSQS with ligand, FSPP | | Descriptor: | Farnesyltransferase, putative, MAGNESIUM ION, ... | | Authors: | Shang, N, Li, Q, Ko, T.P, Chan, H.C, Huang, C.H, Oldfield, E, Guo, R.T. | | Deposit date: | 2013-05-26 | | Release date: | 2014-06-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Squalene synthase as a target for Chagas disease therapeutics.
Plos Pathog., 10, 2014
|
|
3WZL
 
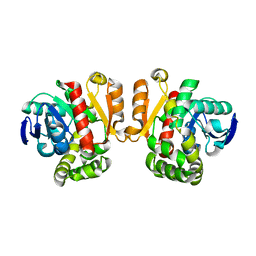 | | ZEN lactonase | | Descriptor: | Zearalenone hydrolase | | Authors: | Ko, T.P, Huang, C.H, Liu, J.R, Guo, R.T. | | Deposit date: | 2014-10-01 | | Release date: | 2014-11-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure and substrate-binding mode of the mycoestrogen-detoxifying lactonase ZHD from Clonostachys rosea
RSC ADV, 4, 2014
|
|
3WZM
 
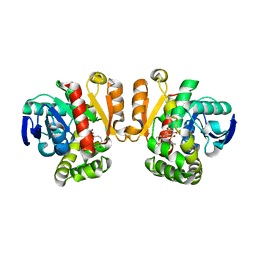 | | ZEN lactonase mutant complex | | Descriptor: | (3S,11E)-14,16-dihydroxy-3-methyl-3,4,5,6,9,10-hexahydro-1H-2-benzoxacyclotetradecine-1,7(8H)-dione, Zearalenone hydrolase | | Authors: | Ko, T.P, Huang, C.H, Liu, J.R, Guo, R.T. | | Deposit date: | 2014-10-01 | | Release date: | 2014-11-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Crystal structure and substrate-binding mode of the mycoestrogen-detoxifying lactonase ZHD from Clonostachys rosea
RSC ADV, 4, 2014
|
|
4FRS
 
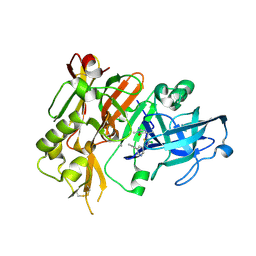 | |
5GTB
 
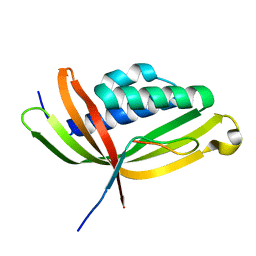 | |
4H3J
 
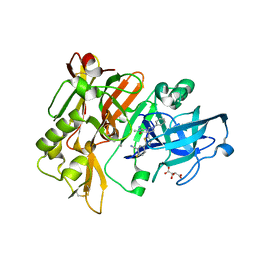 | | Structure of BACE Bound to 2-fluoro-5-(5-(2-imino-3-methyl-4-oxo-6-phenyloctahydro-1H-pyrrolo[3,4-d]pyrimidin-7a-yl)thiophen-2-yl)benzonitrile | | Descriptor: | 2-fluoro-5-{5-[(2E,4aR,7aR)-2-imino-3-methyl-4-oxo-6-phenyloctahydro-7aH-pyrrolo[3,4-d]pyrimidin-7a-yl]thiophen-2-yl}benzonitrile, Beta-secretase 1, L(+)-TARTARIC ACID | | Authors: | Strickland, C, Mandal, M. | | Deposit date: | 2012-09-13 | | Release date: | 2012-10-17 | | Last modified: | 2012-11-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Design and Validation of Bicyclic Iminopyrimidinones As Beta Amyloid Cleaving Enzyme-1 (BACE1) Inhibitors: Conformational Constraint to Favor a Bioactive Conformation.
J.Med.Chem., 55, 2012
|
|
4H3F
 
 | | Structure of BACE Bound to 3-(5-((7aR)-2-imino-6-(6-methoxypyridin-2-yl)-3-methyl-4-oxooctahydro-1H-pyrrolo[3,4-d]pyrimidin-7a-yl)thiophen-3-yl)benzonitrile | | Descriptor: | 3-{5-[(2E,4aR,7aR)-2-imino-6-(6-methoxypyridin-2-yl)-3-methyl-4-oxooctahydro-7aH-pyrrolo[3,4-d]pyrimidin-7a-yl]thiophen-3-yl}benzonitrile, Beta-secretase 1, L(+)-TARTARIC ACID | | Authors: | Strickland, C, Mandal, M. | | Deposit date: | 2012-09-13 | | Release date: | 2012-11-07 | | Last modified: | 2012-11-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design and Validation of Bicyclic Iminopyrimidinones As Beta Amyloid Cleaving Enzyme-1 (BACE1) Inhibitors: Conformational Constraint to Favor a Bioactive Conformation.
J.Med.Chem., 55, 2012
|
|
4H3G
 
 | | Structure of BACE Bound to 2-((7aR)-7a-(4-(3-cyanophenyl)thiophen-2-yl)-2-imino-3-methyl-4-oxohexahydro-1H-pyrrolo[3,4-d]pyrimidin-6(2H)-yl)nicotinonitrile | | Descriptor: | 2-{(2E,4aR,7aR)-7a-[4-(3-cyanophenyl)thiophen-2-yl]-2-imino-3-methyl-4-oxooctahydro-6H-pyrrolo[3,4-d]pyrimidin-6-yl}pyridine-3-carbonitrile, Beta-secretase 1, L(+)-TARTARIC ACID | | Authors: | Strickland, C, Mandal, M. | | Deposit date: | 2012-09-13 | | Release date: | 2012-11-07 | | Last modified: | 2012-11-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Design and Validation of Bicyclic Iminopyrimidinones As Beta Amyloid Cleaving Enzyme-1 (BACE1) Inhibitors: Conformational Constraint to Favor a Bioactive Conformation.
J.Med.Chem., 55, 2012
|
|
4HA5
 
 | | Structure of BACE Bound to (S)-3-(5-(2-imino-1,4-dimethyl-6-oxohexahydropyrimidin-4-yl)thiophen-3-yl)benzonitrile | | Descriptor: | 3-{5-[(2E,4S)-2-imino-1,4-dimethyl-6-oxohexahydropyrimidin-4-yl]thiophen-3-yl}benzonitrile, Beta-secretase 1, L(+)-TARTARIC ACID | | Authors: | Strickland, C, Mandal, M. | | Deposit date: | 2012-09-25 | | Release date: | 2012-10-17 | | Last modified: | 2012-11-21 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Design and Validation of Bicyclic Iminopyrimidinones As Beta Amyloid Cleaving Enzyme-1 (BACE1) Inhibitors: Conformational Constraint to Favor a Bioactive Conformation.
J.Med.Chem., 55, 2012
|
|
5HD0
 
 | |
5HDU
 
 | |
5HDV
 
 | |
4H3I
 
 | | Structure of BACE Bound to 3-(5-((7aR)-2-imino-6-(3-methoxypyridin-2-yl)-3-methyl-4-oxooctahydro-1H-pyrrolo[3,4-d]pyrimidin-7a-yl)thiophen-3-yl)benzonitrile | | Descriptor: | 3-{5-[(2E,4aR,7aR)-2-imino-6-(3-methoxypyridin-2-yl)-3-methyl-4-oxooctahydro-7aH-pyrrolo[3,4-d]pyrimidin-7a-yl]thiophen-3-yl}benzonitrile, Beta-secretase 1, L(+)-TARTARIC ACID | | Authors: | Strickland, C, Mandal, M. | | Deposit date: | 2012-09-13 | | Release date: | 2012-11-07 | | Last modified: | 2012-11-21 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Design and Validation of Bicyclic Iminopyrimidinones As Beta Amyloid Cleaving Enzyme-1 (BACE1) Inhibitors: Conformational Constraint to Favor a Bioactive Conformation.
J.Med.Chem., 55, 2012
|
|
5ZM6
 
 | | Crystal structure of ORP1-ORD in complex with PI(4,5)P2 | | Descriptor: | ACETATE ION, Oxysterol-binding protein-related protein 1, [(2~{S})-1-octadecanoyloxy-3-[oxidanyl-[(1~{R},2~{R},3~{S},4~{S},5~{S},6~{S})-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propan-2-yl] icosa-5,8,11,14-tetraenoate | | Authors: | Dong, J, Wang, J, Luo, Z, Wu, J.W. | | Deposit date: | 2018-04-01 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Allosteric enhancement of ORP1-mediated cholesterol transport by PI(4,5)P2/PI(3,4)P2.
Nat Commun, 10, 2019
|
|
8IMY
 
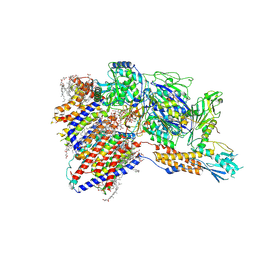 | | Cryo-EM structure of GPI-T (inactive mutant) with GPI and proULBP2, a proprotein substrate | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, T, Xu, Y, Qu, Q, Li, D. | | Deposit date: | 2023-03-07 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Structures of liganded glycosylphosphatidylinositol transamidase illuminate GPI-AP biogenesis.
Nat Commun, 14, 2023
|
|
8IMX
 
 | | Cryo-EM structure of GPI-T with a chimeric GPI-anchored protein | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Xu, Y, Li, T, Qu, Q, Li, D. | | Deposit date: | 2023-03-07 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structures of liganded glycosylphosphatidylinositol transamidase illuminate GPI-AP biogenesis.
Nat Commun, 14, 2023
|
|
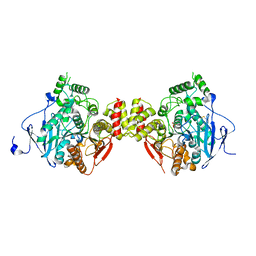
5HCU
 
 | |
5HE7
 
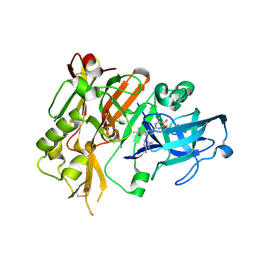 | | BACE-1 in complex with (4aR,7aS)-7a-(2,4-difluorophenyl)-6-(5-fluoro-4-methoxy-6-methylpyrimidin-2-yl)-2-imino-3-methyloctahydro-4H-pyrrolo[3,4-d]pyrimidin-4-one | | Descriptor: | (2E,4aR,7aS)-7a-(2,4-difluorophenyl)-6-(5-fluoro-4-methoxy-6-methylpyrimidin-2-yl)-2-imino-3-methyloctahydro-4H-pyrrolo[3,4-d]pyrimidin-4-one, Beta-secretase 1 | | Authors: | Orth, P. | | Deposit date: | 2016-01-05 | | Release date: | 2016-03-16 | | Last modified: | 2016-04-27 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structure-Based Design of an Iminoheterocyclic beta-Site Amyloid Precursor Protein Cleaving Enzyme (BACE) Inhibitor that Lowers Central A beta in Nonhuman Primates.
J.Med.Chem., 59, 2016
|
|
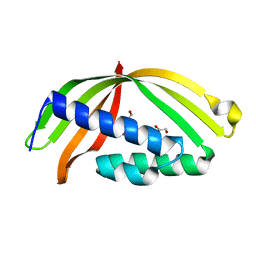
5HAD
 
 | |
