7BQY
 
 | | THE CRYSTAL STRUCTURE OF COVID-19 MAIN PROTEASE IN COMPLEX WITH AN INHIBITOR N3 at 1.7 angstrom | | Descriptor: | 3C-like proteinase, N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE | | Authors: | Liu, X, Zhang, B, Jin, Z, Yang, H, Rao, Z. | | Deposit date: | 2020-03-26 | | Release date: | 2020-04-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of Mprofrom SARS-CoV-2 and discovery of its inhibitors.
Nature, 582, 2020
|
|
6LQ6
 
 | |
6LQ1
 
 | |
6LQ4
 
 | |
6LQ0
 
 | |
5X0X
 
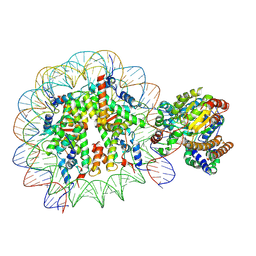 | | Complex of Snf2-Nucleosome complex with Snf2 bound to position +6 of the nucleosome | | Descriptor: | DNA (167-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Li, M, Liu, X, Xia, X, Chen, Z, Li, X. | | Deposit date: | 2017-01-23 | | Release date: | 2017-04-19 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.97 Å) | | Cite: | Mechanism of chromatin remodelling revealed by the Snf2-nucleosome structure.
Nature, 544, 2017
|
|
5X0Y
 
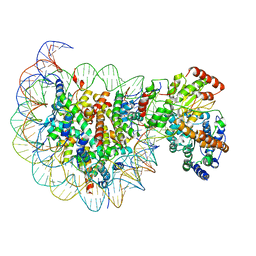 | | Complex of Snf2-Nucleosome complex with Snf2 bound to SHL2 of the nucleosome | | Descriptor: | DNA (167-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Li, M, Liu, X, Xia, X, Chen, Z, Li, X. | | Deposit date: | 2017-01-23 | | Release date: | 2017-04-19 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.69 Å) | | Cite: | Mechanism of chromatin remodelling revealed by the Snf2-nucleosome structure.
Nature, 544, 2017
|
|
6LQ2
 
 | |
3R24
 
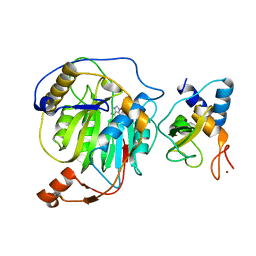 | | Crystal structure of nsp10/nsp16 complex of SARS coronavirus | | Descriptor: | 2'-O-methyl transferase, Non-structural protein 10 and Non-structural protein 11, S-ADENOSYLMETHIONINE, ... | | Authors: | Liu, X, Guo, D, Su, C, Chen, Y. | | Deposit date: | 2011-03-13 | | Release date: | 2011-10-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biochemical and structural insights into the mechanisms of SARS coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex.
Plos Pathog., 7, 2011
|
|
5HP7
 
 | | Crystal structures of RidA in the apo form | | Descriptor: | Reactive Intermediate Deaminase A, chloroplastic | | Authors: | Xie, W, Liu, X. | | Deposit date: | 2016-01-20 | | Release date: | 2016-10-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of RidA, an important enzyme for the prevention of toxic side products
Sci Rep, 6, 2016
|
|
5HP8
 
 | | Crystal structures of RidA in complex with pyruvate | | Descriptor: | PYRUVIC ACID, Reactive Intermediate Deaminase A, chloroplastic | | Authors: | Xie, W, Liu, X. | | Deposit date: | 2016-01-20 | | Release date: | 2016-10-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of RidA, an important enzyme for the prevention of toxic side products
Sci Rep, 6, 2016
|
|
4KFW
 
 | |
4KFV
 
 | |
4QBA
 
 | |
4NXE
 
 | | Crystal structure of iLOV-I486(2LT) at pH 6.5 | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin-2 | | Authors: | Wang, J, Liu, X, Li, J. | | Deposit date: | 2013-12-09 | | Release date: | 2014-09-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | Significant expansion of fluorescent protein sensing ability through the genetic incorporation of superior photo-induced electron-transfer quenchers.
J.Am.Chem.Soc., 136, 2014
|
|
4NXF
 
 | | Crystal structure of iLOV-I486(2LT) at pH 8.0 | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin-2 | | Authors: | Wang, J, Liu, X, Li, J. | | Deposit date: | 2013-12-09 | | Release date: | 2014-09-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.766 Å) | | Cite: | Significant expansion of fluorescent protein sensing ability through the genetic incorporation of superior photo-induced electron-transfer quenchers.
J.Am.Chem.Soc., 136, 2014
|
|
8WEJ
 
 | |
4NXG
 
 | | Crystal structure of iLOV-I486z(2LT) at pH 9.0 | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin-2 | | Authors: | Wang, J, Liu, X, Li, J. | | Deposit date: | 2013-12-09 | | Release date: | 2014-09-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Significant expansion of fluorescent protein sensing ability through the genetic incorporation of superior photo-induced electron-transfer quenchers.
J.Am.Chem.Soc., 136, 2014
|
|
4NXB
 
 | | Crystal structure of iLOV-I486(2LT) at pH 7.0 | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin-2 | | Authors: | Wang, J, Li, J, Liu, X. | | Deposit date: | 2013-12-09 | | Release date: | 2014-09-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.561 Å) | | Cite: | Significant expansion of fluorescent protein sensing ability through the genetic incorporation of superior photo-induced electron-transfer quenchers.
J.Am.Chem.Soc., 136, 2014
|
|
8X2L
 
 | |
2RU9
 
 | |
4GF6
 
 | | crystal structure of GFP with cuprum bound at the Incorporated metal Chelating Amino Acid PYZ151 | | Descriptor: | CALCIUM ION, COPPER (II) ION, green fluorescent protein | | Authors: | Dong, J, Liu, X, Li, J, Wang, J, Gong, W. | | Deposit date: | 2012-08-03 | | Release date: | 2012-08-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Genetic incorporation of a metal-chelating amino Acid as a probe for protein electron transfer.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
4GES
 
 | | crystal structure of GFP-TYR151PYZ with an unnatural amino acid incorporation | | Descriptor: | Green fluorescent protein | | Authors: | Dong, J, Liu, X, Li, J, Wang, J, Gong, W. | | Deposit date: | 2012-08-02 | | Release date: | 2012-08-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Genetic incorporation of a metal-chelating amino Acid as a probe for protein electron transfer.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
4JFG
 
 | | Crystal structure of sfGFP-66-HqAla | | Descriptor: | CESIUM ION, Green fluorescent protein, quinolin-8-ol | | Authors: | Wang, J, Liu, X, Li, J, Zhang, W, Hu, M, Zhou, J. | | Deposit date: | 2013-02-28 | | Release date: | 2013-10-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | Significant expansion of the fluorescent protein chromophore through the genetic incorporation of a metal-chelating unnatural amino acid.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
7DF1
 
 | | Crystal structure of human CD98 heavy chain extracellular domain in complex with S1-F4 scFv | | Descriptor: | 4F2 cell-surface antigen heavy chain, IGL c2062_light_IGKV4-1_IGKJ5, S1-F4 VH | | Authors: | Liu, X, Ding, J, Sui, J, Tian, X. | | Deposit date: | 2020-11-06 | | Release date: | 2022-12-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.806 Å) | | Cite: | An anti-CD98 antibody displaying pH-dependent Fc-mediated tumour-specific activity against multiple cancers in CD98-humanized mice.
Nat Biomed Eng, 2022
|
|
