6A73
 
 | | Complex structure of CSN2 with IP6 | | Descriptor: | COP9 signalosome complex subunit 2,Endolysin, INOSITOL HEXAKISPHOSPHATE, SULFATE ION | | Authors: | Liu, L, Li, D, Rao, F, Wang, T. | | Deposit date: | 2018-07-02 | | Release date: | 2019-07-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.447 Å) | | Cite: | Basis for metabolite-dependent Cullin-RING ligase deneddylation by the COP9 signalosome.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4FYN
 
 | |
7KX5
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with noncovalent inhibitor Jun8-76-3A | | Descriptor: | 3C-like proteinase, GLYCEROL, N-([1,1'-biphenyl]-4-yl)-N-[(1R)-2-oxo-2-{[(1S)-1-phenylethyl]amino}-1-(pyridin-3-yl)ethyl]furan-2-carboxamide | | Authors: | Sacco, M, Wang, J, Chen, Y. | | Deposit date: | 2020-12-03 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of Di- and Trihaloacetamides as Covalent SARS-CoV-2 Main Protease Inhibitors with High Target Specificity.
J.Am.Chem.Soc., 143, 2021
|
|
2MQG
 
 | |
4GQ9
 
 | | Chikungunya virus neutralizing antibody 9.8B Fab fragment | | Descriptor: | Chikungunya virus neutralizing antibody 9.8B Fab fragment heavy chain, Chikungunya virus neutralizing antibody 9.8B Fab fragment light chain | | Authors: | Sun, S, Rossmann, M.G. | | Deposit date: | 2012-08-22 | | Release date: | 2013-07-10 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.995 Å) | | Cite: | Structural analyses at pseudo atomic resolution of Chikungunya virus and antibodies show mechanisms of neutralization.
Elife, 2, 2013
|
|
2N7S
 
 | | Solution Structure of Leptospiral LigA4 Big Domain | | Descriptor: | Ig-like repeat domain protein 1 | | Authors: | Mei, S. | | Deposit date: | 2015-09-17 | | Release date: | 2015-10-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of leptospiral LigA4 Big domain.
Biochem. Biophys. Res. Commun., 467, 2015
|
|
2NB4
 
 | | Solution structure of Q388A3 PDZ domain | | Descriptor: | Putative uncharacterized protein | | Authors: | Mei, S. | | Deposit date: | 2016-01-24 | | Release date: | 2016-02-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Q388A3 PDZ domain from Trypanosoma brucei
J.Struct.Biol., 194, 2016
|
|
3NVI
 
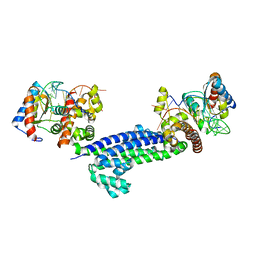 | | Structure of N-terminal truncated Nop56/58 bound with L7Ae and box C/D RNA | | Descriptor: | 50S ribosomal protein L7Ae, NOP5/NOP56 related protein, RNA (5'-R(*CP*UP*CP*UP*GP*AP*CP*CP*GP*AP*AP*AP*GP*GP*CP*GP*UP*GP*AP*UP*GP*AP*GP*C)-3') | | Authors: | Li, H, Xue, S, Wang, R. | | Deposit date: | 2010-07-08 | | Release date: | 2011-07-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.709 Å) | | Cite: | Structural basis for substrate placement by an archaeal box C/D ribonucleoprotein particle.
Mol.Cell, 39, 2010
|
|
4HRG
 
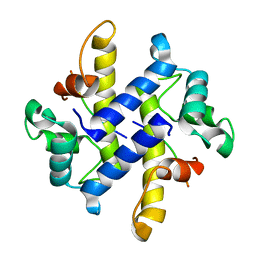 | |
4I0R
 
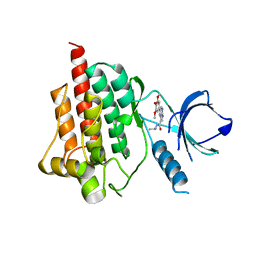 | | Crystal structure of spleen tyrosine kinase complexed with 2-(3,4,5-Trimethoxy-phenyl)-5H-pyrrolo[2,3-b]pyrazine-7-carboxylic acid isopropylamide | | Descriptor: | N-(propan-2-yl)-2-(3,4,5-trimethoxyphenyl)-5H-pyrrolo[2,3-b]pyrazine-7-carboxamide, Tyrosine-protein kinase SYK | | Authors: | Kuglstatter, A, Villasenor, A.G. | | Deposit date: | 2012-11-19 | | Release date: | 2013-10-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Pyrrolopyrazines as selective spleen tyrosine kinase inhibitors.
J.Med.Chem., 56, 2013
|
|
4LX4
 
 | | Crystal Structure Determination of Pseudomonas stutzeri endoglucanase Cel5A using a Twinned Data Set | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Endoglucanase(Endo-1,4-beta-glucanase)protein | | Authors: | Dutoit, R, Delsaute, M, Berlemont, R, Van Elder, D, Galleni, M, Bauvois, C. | | Deposit date: | 2013-07-29 | | Release date: | 2014-07-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.556 Å) | | Cite: | Crystal structure determination of Pseudomonas stutzeri A1501 endoglucanase Cel5A: the search for a molecular basis for glycosynthesis in GH5_5 enzymes.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
4I0T
 
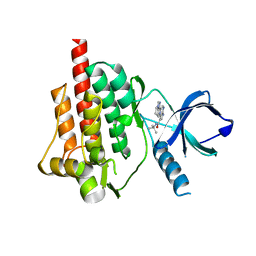 | | Crystal structure of spleen tyrosine kinase complexed with 2-(5,6,7,8-Tetrahydro-imidazo[1,5-a]pyridin-1-yl)-5H-pyrrolo[2,3-b]pyrazine-7-carboxylic acid tert-butylamide | | Descriptor: | N-tert-butyl-2-(5,6,7,8-tetrahydroimidazo[1,5-a]pyridin-1-yl)-5H-pyrrolo[2,3-b]pyrazine-7-carboxamide, Tyrosine-protein kinase SYK | | Authors: | Kuglstatter, A, Slade, M. | | Deposit date: | 2012-11-19 | | Release date: | 2013-10-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Pyrrolopyrazines as selective spleen tyrosine kinase inhibitors.
J.Med.Chem., 56, 2013
|
|
3PB1
 
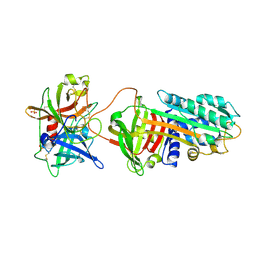 | | Crystal Structure of a Michaelis Complex between Plasminogen Activator Inhibitor-1 and Urokinase-type Plasminogen Activator | | Descriptor: | Plasminogen activator inhibitor 1, Plasminogen activator, urokinase, ... | | Authors: | Lin, Z, Jiang, L, Huang, M, Structure 2 Function Project (S2F) | | Deposit date: | 2010-10-20 | | Release date: | 2010-12-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for recognition of urokinase-type plasminogen activator by plasminogen activator inhibitor-1.
J.Biol.Chem., 286, 2011
|
|
3P8B
 
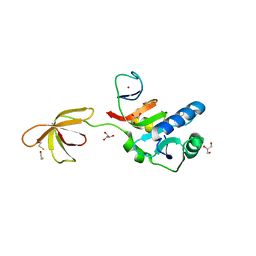 | |
3QE7
 
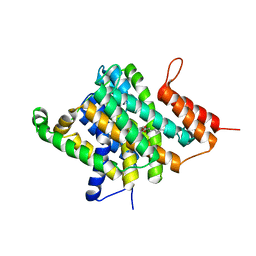 | | Crystal Structure of Uracil Transporter--UraA | | Descriptor: | URACIL, Uracil permease, nonyl beta-D-glucopyranoside | | Authors: | Lu, F.R, Li, S, Yan, N. | | Deposit date: | 2011-01-20 | | Release date: | 2011-03-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.781 Å) | | Cite: | Structure and mechanism of the uracil transporter UraA
Nature, 472, 2011
|
|
7RFK
 
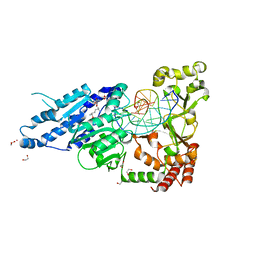 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Inhibitor Sinefungin | | Descriptor: | 1,2-ETHANEDIOL, DNA Strand 1, DNA Strand 2, ... | | Authors: | Horton, J.R, Cheng, X, Zhou, J. | | Deposit date: | 2021-07-14 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Repurposing epigenetic inhibitors to target the Clostridioides difficile- specific DNA adenine methyltransferase and sporulation regulator CamA.
Epigenetics, 17, 2022
|
|
7RFM
 
 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Inhibitor EPZ004777 | | Descriptor: | 1,2-ETHANEDIOL, 7-{5-[(3-{[(4-tert-butylphenyl)carbamoyl]amino}propyl)(propan-2-yl)amino]-5-deoxy-beta-D-ribofuranosyl}-7H-pyrrolo[2,3-d]pyrimidin-4-amine, DNA Strand 1, ... | | Authors: | Horton, J.R, Cheng, X, Zhou, J. | | Deposit date: | 2021-07-14 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Repurposing epigenetic inhibitors to target the Clostridioides difficile- specific DNA adenine methyltransferase and sporulation regulator CamA.
Epigenetics, 17, 2022
|
|
7RFN
 
 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Inhibitor SGC8158 | | Descriptor: | 1,2-ETHANEDIOL, 5'-S-(4-{[(4'-chloro[1,1'-biphenyl]-3-yl)methyl]amino}butyl)-5'-thioadenosine, DNA Strand 1, ... | | Authors: | Horton, J.R, Cheng, X, Zhou, J. | | Deposit date: | 2021-07-14 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Repurposing epigenetic inhibitors to target the Clostridioides difficile- specific DNA adenine methyltransferase and sporulation regulator CamA.
Epigenetics, 17, 2022
|
|
7RFL
 
 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Inhibitor SGC0946 | | Descriptor: | 1,2-ETHANEDIOL, 5-bromo-7-{5-[(3-{[(4-tert-butylphenyl)carbamoyl]amino}propyl)(propan-2-yl)amino]-5-deoxy-beta-D-ribofuranosyl}-7H-pyrrolo[2,3-d]pyrimidin-4-amine, DNA Strand 1, ... | | Authors: | Horton, J.R, Cheng, X, Zhou, J. | | Deposit date: | 2021-07-14 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Repurposing epigenetic inhibitors to target the Clostridioides difficile- specific DNA adenine methyltransferase and sporulation regulator CamA.
Epigenetics, 17, 2022
|
|
4HJE
 
 | | Crystal structure of p53 core domain in complex with DNA | | Descriptor: | Cellular tumor antigen p53, DNA (5'-D(*AP*GP*GP*CP*TP*TP*GP*TP*CP*TP*CP*TP*AP*AP*CP*TP*TP*GP*TP*GP*A)-3'), DNA (5'-D(*TP*CP*AP*CP*AP*AP*GP*TP*TP*AP*GP*AP*GP*AP*CP*AP*AP*GP*CP*CP*T)-3'), ... | | Authors: | Chen, Y, Chen, L. | | Deposit date: | 2012-10-12 | | Release date: | 2013-07-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.907 Å) | | Cite: | Structure of p53 binding to the BAX response element reveals DNA unwinding and compression to accommodate base-pair insertion.
Nucleic Acids Res., 41, 2013
|
|
9E6R
 
 | |
9E6S
 
 | |
9E6T
 
 | |
9JMZ
 
 | | Cryo-EM structure of a human-infecting bovine influenza H5N1 hemagglutinin complexed with avian receptor analog LSTa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Wang, H.C, Han, P, Song, H, Gao, G.F. | | Deposit date: | 2024-09-22 | | Release date: | 2025-01-22 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Receptor binding, structure, and tissue tropism of cattle-infecting H5N1 avian influenza virus hemagglutinin.
Cell, 188, 2025
|
|
8XC4
 
 | | Nipah virus attachment glycoprotein head domain in complex with a broadly neutralizing antibody 1E5 | | Descriptor: | 1E5-VH, 1E5-VL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Fan, P.F, Yu, C.M, Chen, W. | | Deposit date: | 2023-12-08 | | Release date: | 2024-01-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | A potent Henipavirus cross-neutralizing antibody reveals a dynamic fusion-triggering pattern of the G-tetramer.
Nat Commun, 15, 2024
|
|
