2DRK
 
 | |
4G1R
 
 | | Crystal structure of anti-HIV actinohivin in complex with alphs-1,2-mannobiose (Form II) | | Descriptor: | Actinohivin, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose | | Authors: | Hoque, M.M, Suzuki, K, Tsunoda, M, Jiang, J, Zhang, F, Takahashi, A, Naomi, O, Zhang, X, Sekiguchi, T, Tanaka, H, Omura, S, Takenaka, A. | | Deposit date: | 2012-07-11 | | Release date: | 2013-07-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Matured structure of anti-HIV lectin actinohivin in complex with alpha-1,2-mannobiose
To be Published
|
|
1WZL
 
 | | Thermoactinomyces vulgaris R-47 alpha-amylase II (TVA II) mutatnt R469L | | Descriptor: | Alpha-amylase II, CALCIUM ION | | Authors: | Mizuno, M, Ichikawa, K, Tonozuka, T, Ohtaki, A, Shimura, Y, Kamitori, S, Nishikawa, A, Sakano, Y. | | Deposit date: | 2005-03-06 | | Release date: | 2005-03-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mutagenesis and Structural Analysis of Thermoactinomyces vulgaris R-47 alpha-Amylase II (TVA II)
To be Published
|
|
4GAA
 
 | | Structure of Leukotriene A4 hydrolase from Xenopus laevis complexed with inhibitor bestatin | | Descriptor: | 2-(3-AMINO-2-HYDROXY-4-PHENYL-BUTYRYLAMINO)-4-METHYL-PENTANOIC ACID, MGC78867 protein, ZINC ION | | Authors: | Stsiapanava, A, Kumar, R.B, Haeggstrom, J.Z, Rinaldo-Matthis, A. | | Deposit date: | 2012-07-25 | | Release date: | 2013-06-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Product formation controlled by substrate dynamics in leukotriene A4 hydrolase.
Biochim.Biophys.Acta, 1844, 2014
|
|
6RKO
 
 | | Cryo-EM structure of the E. coli cytochrome bd-I oxidase at 2.68 A resolution | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, Cytochrome bd-I ubiquinol oxidase subunit 1, ... | | Authors: | Safarian, S, Hahn, A, Kuehlbrandt, W, Michel, H. | | Deposit date: | 2019-04-30 | | Release date: | 2019-10-16 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Active site rearrangement and structural divergence in prokaryotic respiratory oxidases.
Science, 366, 2019
|
|
6RGZ
 
 | | Revisiting pH-gated conformational switch. Complex HK853-RR468 pH 6.5 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Mideros-Mora, C, Casino, P, Marina, A. | | Deposit date: | 2019-04-18 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Revisiting the pH-gated conformational switch on the activities of HisKA-family histidine kinases.
Nat Commun, 11, 2020
|
|
1INW
 
 | | A SIALIC ACID DERIVED PHOSPHONATE ANALOG INHIBITS DIFFERENT STRAINS OF INFLUENZA VIRUS NEURAMINIDASE WITH DIFFERENT EFFICIENCIES | | Descriptor: | (1S)-4-acetamido-1,5-anhydro-2,4-dideoxy-1-phosphono-D-glycero-D-galacto-octitol, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | White, C.L, Janakiraman, M.N, Laver, W.G, Philippon, C, Vasella, A, Air, G.M, Luo, M. | | Deposit date: | 1994-09-26 | | Release date: | 1995-02-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A sialic acid-derived phosphonate analog inhibits different strains of influenza virus neuraminidase with different efficiencies.
J.Mol.Biol., 245, 1995
|
|
6RH8
 
 | | Revisiting pH-gated conformational switch. Complex HK853 mutant H260A -RR468 mutant D53A pH 5.3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Response regulator, SULFATE ION, ... | | Authors: | Mideros-Mora, C, Casino, P, Marina, A. | | Deposit date: | 2019-04-19 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Revisiting the pH-gated conformational switch on the activities of HisKA-family histidine kinases.
Nat Commun, 11, 2020
|
|
3VU9
 
 | | Crystal Structure of Psy3-Csm2 complex | | Descriptor: | 1,2-ETHANEDIOL, Chromosome segregation in meiosis protein 2, Platinum sensitivity protein 3 | | Authors: | Tawaramoto, M, Sasanuma, H, Hosaka, H, Lao, J.P, Sanda, E, Suzuki, M, Yamashita, E, Hunter, N, Shinohara, M, Nakagawa, A, Shinohara, A. | | Deposit date: | 2012-06-23 | | Release date: | 2013-04-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A new protein complex promoting the assembly of Rad51 filaments
Nat Commun, 4, 2013
|
|
3RMN
 
 | | Human Thrombin in complex with MI341 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Hirudin variant-2, ... | | Authors: | Biela, A, Heine, A, Klebe, G. | | Deposit date: | 2011-04-21 | | Release date: | 2012-04-25 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Ligand binding stepwise disrupts water network in thrombin: enthalpic and entropic changes reveal classical hydrophobic effect
J.Med.Chem., 55, 2012
|
|
8F76
 
 | | Human olfactory receptor OR51E2 bound to propionate in complex with miniGs399 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Billesboelle, C.B, Manglik, A. | | Deposit date: | 2022-11-18 | | Release date: | 2023-03-22 | | Last modified: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of odorant recognition by a human odorant receptor.
Nature, 615, 2023
|
|
3IAR
 
 | | The crystal structure of human adenosine deaminase | | Descriptor: | (2R,3S,5R)-5-(6-amino-9H-purin-9-yl)-tetrahydro-2-(hydroxymethyl)furan-3-ol, Adenosine deaminase, GLYCEROL, ... | | Authors: | Ugochukwu, E, Zhang, Y, Hapka, E, Yue, W.W, Bray, J.E, Muniz, J, Burgess-Brown, N, Chaikuad, A, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Kavanagh, K.L, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-07-14 | | Release date: | 2009-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | The crystal structure of human adenosine deaminase
To be Published
|
|
7NET
 
 | |
7NES
 
 | |
7NER
 
 | |
6RNN
 
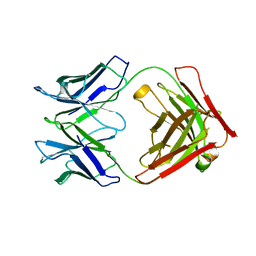 | | P46, an immunodominant surface protein from Mycoplasma hyopneumoniae | | Descriptor: | Immunoglobulin heavy chain, Immunoglobulin light chain | | Authors: | Guasch, A, Gonzalez-Gonzalez, L, Fita, I. | | Deposit date: | 2019-05-09 | | Release date: | 2019-11-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of P46, an immunodominant surface protein from Mycoplasma hyopneumoniae: interaction with a monoclonal antibody.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
3R4U
 
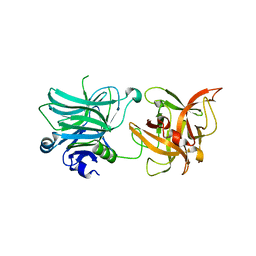 | | Cell entry of botulinum neurotoxin type C is dependent upon interaction with two ganglioside molecules | | Descriptor: | Botulinum neurotoxin type C1 | | Authors: | Strotmeier, J, Gu, S, Jutzi, S, Mahrhold, S, Zhou, J, Pich, A, Bigalke, H, Rummel, A, Jin, R, Binz, T. | | Deposit date: | 2011-03-17 | | Release date: | 2011-06-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The biological activity of botulinum neurotoxin type C is dependent upon novel types of ganglioside binding sites.
Mol.Microbiol., 81, 2011
|
|
3VK9
 
 | | Crystal structure of delta-class glutathione transferase from silkmoth | | Descriptor: | GLYCEROL, Glutathione S-transferase delta | | Authors: | Kakuta, Y, Usuda, K, Higashiura, A, Suzuki, M, Nakagawa, A, Kimura, M, Yamamoto, K. | | Deposit date: | 2011-11-10 | | Release date: | 2012-10-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural basis for catalytic activity of a silkworm Delta-class glutathione transferase
Biochim.Biophys.Acta, 1820, 2012
|
|
1WPN
 
 | | Crystal structure of the N-terminal core of Bacillus subtilis inorganic pyrophosphatase | | Descriptor: | MANGANESE (II) ION, Manganese-dependent inorganic pyrophosphatase, SULFATE ION | | Authors: | Fabrichniy, I.P, Lehtio, L, Salminen, A, Baykov, A.A, Lahti, R, Goldman, A. | | Deposit date: | 2004-09-09 | | Release date: | 2004-11-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural Studies of Metal Ions in Family II Pyrophosphatases: The Requirement for a Janus Ion
Biochemistry, 43, 2004
|
|
2MUC
 
 | | MUCONATE CYCLOISOMERASE VARIANT F329I | | Descriptor: | MANGANESE (II) ION, PROTEIN (MUCONATE CYCLOISOMERASE) | | Authors: | Schell, U, Helin, S, Kajander, T, Schlomann, M, Goldman, A. | | Deposit date: | 1998-10-26 | | Release date: | 1999-12-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the activity of two muconate cycloisomerase variants toward substituted muconates.
Proteins, 34, 1999
|
|
3ICR
 
 | | Crystal structure of oxidized Bacillus anthracis CoADR-RHD | | Descriptor: | COENZYME A, Coenzyme A-Disulfide Reductase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Wallen, J.R, Claiborne, A. | | Deposit date: | 2009-07-18 | | Release date: | 2009-11-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure and catalytic properties of Bacillus anthracis CoADR-RHD: implications for flavin-linked sulfur trafficking.
Biochemistry, 48, 2009
|
|
3VSR
 
 | | Microbacterium saccharophilum K-1 beta-fructofuranosidase catalytic domain | | Descriptor: | Beta-fructofuranosidase | | Authors: | Tonozuka, T, Tamaki, A, Yokoi, G, Miyazaki, T, Ichikawa, M, Nishikawa, A, Ohta, Y, Hidaka, Y, Katayama, K, Hatada, Y, Ito, T, Fujita, K. | | Deposit date: | 2012-05-08 | | Release date: | 2012-08-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a lactosucrose-producing enzyme, Arthrobacter sp. K-1 beta-fructofuranosidase
Enzyme.Microb.Technol., 51, 2012
|
|
6S5Y
 
 | |
2MXG
 
 | |
3RMP
 
 | | Structural basis for the recognition of attP substrates by P4-like integrases | | Descriptor: | 5'-D(*TP*AP*AP*TP*GP*AP*CP*CP*AP*CP*CP*AP*AP*TP*A)-3', 5'-D(*TP*AP*TP*TP*GP*GP*TP*GP*GP*TP*CP*AP*TP*TP*A)-3', CP4-like integrase | | Authors: | Szwagierczak, A, Popowicz, G.M, Holak, T.A, Rakin, A, Antonenka, U. | | Deposit date: | 2011-04-21 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural basis for the recognition of attP substrates by P4-like integrases
To be Published
|
|
