8PCQ
 
 | | Structure of serine-beta-lactamase CTX-M-14 following the time-resolved active site binding of boric acid and subsequent glycerol-boric acid-ester formation, 500 ms | | Descriptor: | BORATE ION, Beta-lactamase, SULFATE ION, ... | | Authors: | Prester, A, Perbandt, M, Galchenkova, M, Oberthuer, D, Yefanov, O, Hinrichs, W, Rohde, H, Betzel, C. | | Deposit date: | 2023-06-11 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Time-resolved crystallography of boric acid binding to the active site serine of the beta-lactamase CTX-M-14 and subsequent 1,2-diol esterification.
Commun Chem, 7, 2024
|
|
4WEV
 
 | | Crystal structure of human AKR1B10 complexed with NADP+ and sulindac | | Descriptor: | Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, [(1Z)-5-fluoro-2-methyl-1-{4-[methylsulfinyl]benzylidene}-1H-inden-3-yl]acetic acid | | Authors: | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Crespo, I, Porte, S, Pares, X, Farres, J, Podjarny, A. | | Deposit date: | 2014-09-11 | | Release date: | 2015-01-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.453 Å) | | Cite: | Structural analysis of sulindac as an inhibitor of aldose reductase and AKR1B10.
Chem.Biol.Interact., 234, 2015
|
|
8PCB
 
 | | Structure of serine-beta-lactamase CTX-M-14 following the time-resolved active site binding of boric acid, 80 ms | | Descriptor: | BORATE ION, Beta-lactamase, SULFATE ION | | Authors: | Prester, A, Perbandt, M, Galchenkova, M, Oberthuer, D, Yefanov, O, Hinrichs, W, Rohde, H, Betzel, C. | | Deposit date: | 2023-06-11 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Time-resolved crystallography of boric acid binding to the active site serine of the beta-lactamase CTX-M-14 and subsequent 1,2-diol esterification.
Commun Chem, 7, 2024
|
|
8PCO
 
 | | Structure of serine-beta-lactamase CTX-M-14 following the time-resolved active site binding of boric acid and subsequent glycerol-boric acid-ester formation, 150 ms | | Descriptor: | BORATE ION, Beta-lactamase, SULFATE ION, ... | | Authors: | Prester, A, Perbandt, M, Galchenkova, M, Oberthuer, D, Yefanov, O, Hinrichs, W, Rohde, H, Betzel, C. | | Deposit date: | 2023-06-11 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Time-resolved crystallography of boric acid binding to the active site serine of the beta-lactamase CTX-M-14 and subsequent 1,2-diol esterification.
Commun Chem, 7, 2024
|
|
4WKA
 
 | | Crystal structure of human chitotriosidase-1 catalytic domain at 0.95 A resolution | | Descriptor: | Chitotriosidase-1, L(+)-TARTARIC ACID | | Authors: | Fadel, F, Zhao, Y, Cachau, R, Cousido-Siah, A, Ruiz, F.X, Harlos, K, Howard, E, Mitschler, A, Podjarny, A. | | Deposit date: | 2014-10-02 | | Release date: | 2015-07-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | New insights into the enzymatic mechanism of human chitotriosidase (CHIT1) catalytic domain by atomic resolution X-ray diffraction and hybrid QM/MM.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4WKF
 
 | | Crystal structure of human chitotriosidase-1 catalytic domain in complex with chitobiose (2.5mM) at 1.10 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitotriosidase-1 | | Authors: | Fadel, F, Zhao, Y, Cachau, R, Cousido-Siah, A, Ruiz, F.X, Harlos, K, Howard, E, Mitschler, A, Podjarny, A. | | Deposit date: | 2014-10-02 | | Release date: | 2015-07-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.101 Å) | | Cite: | New insights into the enzymatic mechanism of human chitotriosidase (CHIT1) catalytic domain by atomic resolution X-ray diffraction and hybrid QM/MM.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4X71
 
 | | Crystal Structure of lipase from Geobacillus stearothermophilus T6 methanol stable variant A269T | | Descriptor: | CALCIUM ION, Lipase, ZINC ION | | Authors: | Kanteev, M, Dror, A, Gihaz, S, Shahar, A, Fishman, A. | | Deposit date: | 2014-12-09 | | Release date: | 2015-06-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into methanol-stable variants of lipase T6 from Geobacillus stearothermophilus.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
4WRP
 
 | | The C-terminal domain of gene product lpg0944 from Legionella pneumophila subsp. pneumophila str. Philadelphia 1 | | Descriptor: | Uncharacterized protein | | Authors: | Cuff, M.E, Evdokimova, E, Egorova, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-10-24 | | Release date: | 2015-03-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The C-terminal domain of gene product lpg0944 from Legionella pneumophila subsp. pneumophila str. Philadelphia 1.
To Be Published
|
|
4V62
 
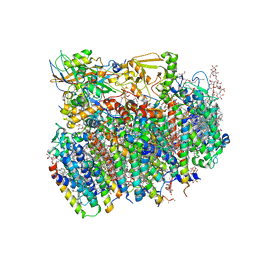 | | Crystal Structure of cyanobacterial Photosystem II | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Guskov, A, Gabdulkhakov, A, Kern, J, Broser, M, Zouni, A, Saenger, W. | | Deposit date: | 2008-01-17 | | Release date: | 2014-07-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Cyanobacterial photosystem II at 2.9-A resolution and the role of quinones, lipids, channels and chloride
Nat.Struct.Mol.Biol., 16, 2009
|
|
4UTB
 
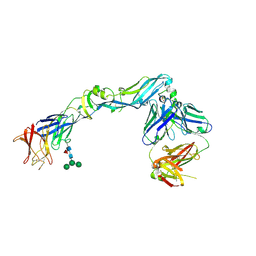 | | Crystal structure of dengue 2 virus envelope glycoprotein in complex with the Fab fragment of the broadly neutralizing human antibody EDE2 A11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BROADLY NEUTRALIZING HUMAN ANTIBODY EDE2 A11, ... | | Authors: | Rouvinski, A, Guardado-Calvo, P, Barba-Spaeth, G, Duquerroy, S, Vaney, M.C, Rey, F.A. | | Deposit date: | 2014-07-18 | | Release date: | 2015-01-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.85 Å) | | Cite: | Recognition Determinants of Broadly Neutralizing Human Antibodies Against Dengue Viruses.
Nature, 520, 2015
|
|
4UT7
 
 | | CRYSTAL STRUCTURE OF THE SCFV FRAGMENT OF THE BROADLY NEUTRALIZING HUMAN ANTIBODY EDE2 A11 | | Descriptor: | BROADLY NEUTRALIZING HUMAN ANTIBODY EDE2 A11 | | Authors: | Rouvinski, A, Guardado-Calvo, P, Barba-Spaeth, G, Duquerroy, S, Vaney, M.C, Rey, F.A. | | Deposit date: | 2014-07-18 | | Release date: | 2015-01-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Recognition Determinants of Broadly Neutralizing Human Antibodies Against Dengue Viruses.
Nature, 520, 2015
|
|
4X6U
 
 | | Crystal Structure of lipase from Geobacillus stearothermophilus T6 | | Descriptor: | CALCIUM ION, Lipase, ZINC ION | | Authors: | Kanteev, M, Dror, A, Gihaz, S, Fishman, A. | | Deposit date: | 2014-12-09 | | Release date: | 2015-06-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Structural insights into methanol-stable variants of lipase T6 from Geobacillus stearothermophilus.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
4X7B
 
 | | Crystal Structure of lipase from Geobacillus stearothermophilus T6 methanol stable variant H86Y/A269T | | Descriptor: | CALCIUM ION, Lipase, ZINC ION | | Authors: | Kanteev, M, Dror, A, Gihaz, S, Fishman, A. | | Deposit date: | 2014-12-09 | | Release date: | 2015-06-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into methanol-stable variants of lipase T6 from Geobacillus stearothermophilus.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
4X85
 
 | | Crystal Structure of lipase from Geobacillus stearothermophilus T6 methanol stable variant H86Y/A269T/R374W | | Descriptor: | CALCIUM ION, Lipase, ZINC ION | | Authors: | Kanteev, M, Dror, A, Gihaz, S, Fishman, A. | | Deposit date: | 2014-12-10 | | Release date: | 2015-06-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.192 Å) | | Cite: | Structural insights into methanol-stable variants of lipase T6 from Geobacillus stearothermophilus.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
7SHQ
 
 | | Structure of a functional construct of eukaryotic elongation factor 2 kinase in complex with calmodulin. | | Descriptor: | CALCIUM ION, Calmodulin-1, Eukaryotic elongation factor 2 kinase,Eukaryotic elongation factor 2 kinase, ... | | Authors: | Piserchio, A, Isiorho, E.A, Jeruzalmi, D, Dalby, K.N, Ghose, R. | | Deposit date: | 2021-10-11 | | Release date: | 2022-04-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural basis for the calmodulin-mediated activation of eukaryotic elongation factor 2 kinase.
Sci Adv, 8, 2022
|
|
7B38
 
 | | Torpedo californica acetylcholinesterase complexed with Mg+2 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase, ... | | Authors: | Silman, I, Shnyrov, V.L, Ashani, Y, Roth, E, Nicolas, A, Sussman, J.L, Weiner, L. | | Deposit date: | 2020-11-29 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Torpedo californica acetylcholinesterase is stabilized by binding of a divalent metal ion to a novel and versatile 4D motif.
Protein Sci., 30, 2021
|
|
7B2W
 
 | | Torpedo californica acetylcholinesterase complexed with UO2 | | Descriptor: | Acetylcholinesterase, URANYL (VI) ION | | Authors: | Silman, I, Shnyrov, V.L, Ashani, Y, Roth, E, Nicolas, A, Sussman, J.L, Weiner, L. | | Deposit date: | 2020-11-28 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Torpedo californica acetylcholinesterase is stabilized by binding of a divalent metal ion to a novel and versatile 4D motif.
Protein Sci., 30, 2021
|
|
6FC9
 
 | | The 1,8-bis(aminomethyl)anthracene and Quadruplex-duplex junction complex | | Descriptor: | DNA (27-MER), [8-(azaniumylmethyl)anthracen-1-yl]methylazanium | | Authors: | Santana, A, Serrano, I, Montalvillo-Jimenez, L, Corzana, F, Bastida, A, Jimenez-Barbero, J, Gonzalez, C, Asensio, J.L. | | Deposit date: | 2017-12-20 | | Release date: | 2019-04-10 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | De Novo Design of Selective Quadruplex-Duplex Junction Ligands and Structural Characterisation of Their Binding Mode: Targeting the G4 Hot-Spot.
Chemistry, 2020
|
|
7AT5
 
 | | Structure of protein kinase ck2 catalytic subunit (csnk2a1 gene product) in complex with the bivalent inhibitor KN2 | | Descriptor: | 1,2-ETHANEDIOL, 2-(3,4-dichlorophenyl)ethanamine, Casein kinase II subunit alpha, ... | | Authors: | Lindenblatt, D, Applegate, V, Nickelsen, A, Klussmann, M, Neundorf, I, Goetz, C, Jose, J, Niefind, K. | | Deposit date: | 2020-10-29 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Molecular Plasticity of Crystalline CK2 alpha ' Leads to KN2, a Bivalent Inhibitor of Protein Kinase CK2 with Extraordinary Selectivity.
J.Med.Chem., 65, 2022
|
|
7ATV
 
 | | Structure of protein kinase ck2 catalytic subunit (csnk2a2 gene product) in complex with the bivalent inhibitor KN2 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Casein kinase II subunit alpha', ... | | Authors: | Lindenblatt, D, Applegate, V, Nickelsen, A, Klussmann, M, Neundorf, I, Goetz, C, Jose, J, Niefind, K. | | Deposit date: | 2020-10-31 | | Release date: | 2021-08-04 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Molecular Plasticity of Crystalline CK2 alpha ' Leads to KN2, a Bivalent Inhibitor of Protein Kinase CK2 with Extraordinary Selectivity.
J.Med.Chem., 65, 2022
|
|
7AT9
 
 | | Structure of protein kinase ck2 catalytic subunit (csnk2a2 gene product) in complex with the ATP-competitive inhibitor MB002 and the alphaD-pocket ligand 3,4-dichlorophenethylamine | | Descriptor: | 1,2-ETHANEDIOL, 2-(3,4-dichlorophenyl)ethanamine, 3-(4,5,6,7-tetrabromo-1H-benzotriazol-1-yl)propan-1-ol, ... | | Authors: | Lindenblatt, D, Applegate, V, Nickelsen, A, Klussmann, M, Neundorf, I, Goetz, C, Jose, J, Niefind, K. | | Deposit date: | 2020-10-29 | | Release date: | 2021-08-04 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Molecular Plasticity of Crystalline CK2 alpha ' Leads to KN2, a Bivalent Inhibitor of Protein Kinase CK2 with Extraordinary Selectivity.
J.Med.Chem., 65, 2022
|
|
7TEH
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (3CL Mpro) in complex with BBH-2 | | Descriptor: | (1R,2S,5S)-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-N-{(1Z,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Kovalevsky, A, Kneller, D.W, Coates, L. | | Deposit date: | 2022-01-05 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Covalent narlaprevir- and boceprevir-derived hybrid inhibitors of SARS-CoV-2 main protease
Nat Commun, 13, 2022
|
|
7TFR
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (3CL Mpro) in complex with NBH-2 | | Descriptor: | (1R,2S,5S)-N-{(1Z,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-({1-[(2-methylpropane-2-sulfonyl)methyl]cyclohexyl}carbamoyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Kovalevsky, A, Kneller, D.W, Coates, L. | | Deposit date: | 2022-01-07 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Covalent narlaprevir- and boceprevir-derived hybrid inhibitors of SARS-CoV-2 main protease
Nat Commun, 13, 2022
|
|
6VV0
 
 | | Crystal structure of Eis from Mycobacterium tuberculosis in complex with inhibitor SGT1354 | | Descriptor: | 2-[(4-amino-6,7-dihydro-5H-cyclopenta[4,5]thieno[2,3-d]pyrimidin-2-yl)sulfanyl]-N-[2-(diethylamino)ethyl]acetamide, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Punetha, A, Hou, C, Ngo, H.X, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2020-02-16 | | Release date: | 2020-06-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure-Guided Optimization of Inhibitors of Acetyltransferase Eis fromMycobacterium tuberculosis.
Acs Chem.Biol., 15, 2020
|
|
1ZUA
 
 | | Crystal Structure Of AKR1B10 Complexed With NADP+ And Tolrestat | | Descriptor: | Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, TOLRESTAT | | Authors: | Gallego, O, Ruiz, F.X, Ardevol, A, Dominguez, M, Alvarez, R, de Lera, A.R, Rovira, C, Farres, J, Fita, I, Pares, X. | | Deposit date: | 2005-05-30 | | Release date: | 2006-11-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural basis for the high all-trans-retinaldehyde reductase activity of the tumor marker AKR1B10.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
