4FGM
 
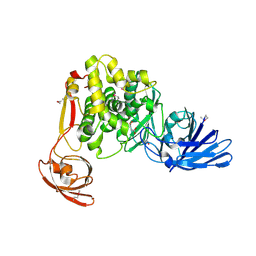 | | Crystal structure of the aminopeptidase N family protein Q5QTY1 from Idiomarina loihiensis. Northeast Structural Genomics Consortium Target IlR60. | | Descriptor: | Aminopeptidase N family protein, MALEIC ACID, ZINC ION | | Authors: | Vorobiev, S, Su, M, Tong, T, Kohan, E, Wang, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-06-04 | | Release date: | 2012-08-01 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.394 Å) | | Cite: | Crystal structure of the aminopeptidase N family protein Q5QTY1 from Idiomarina loihiensis.
To be Published
|
|
3HXJ
 
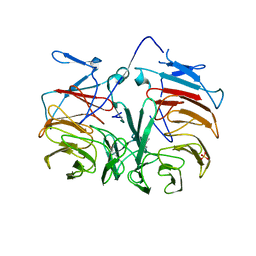 | | Crystal Structure of Pyrrolo-quinoline quinone (PQQ_DH) from Methanococcus maripaludis, Northeast Structural Genomics Consortium Target MrR86 | | Descriptor: | Pyrrolo-quinoline quinone, SULFATE ION | | Authors: | Forouhar, F, Chen, Y, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Foote, E.L, Zhao, L, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-06-20 | | Release date: | 2009-08-25 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Northeast Structural Genomics Consortium Target MrR86
To be Published
|
|
4N6C
 
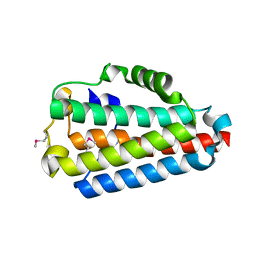 | | Crystal Structure of the B1RZQ2 protein from Streptococcus pneumoniae. Northeast Structural Genomics Consortium (NESG) Target SpR36. | | Descriptor: | BROMIDE ION, uncharacterized protein | | Authors: | Vorobiev, S, Seetharaman, J, Patel, D, Xiao, R, Ciccosanti, C, Wang, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-10-11 | | Release date: | 2013-10-30 | | Method: | X-RAY DIFFRACTION (1.548 Å) | | Cite: | Crystal Structure of the B1RZQ2 protein from Streptococcus pneumoniae.
To be Published
|
|
4F2V
 
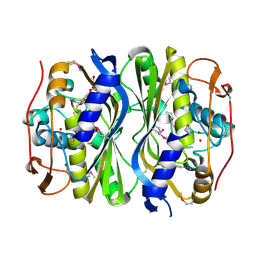 | | Crystal Structure of de novo designed serine hydrolase, Northeast Structural Genomics Consortium (NESG) Target OR165 | | Descriptor: | DI(HYDROXYETHYL)ETHER, DODECYL-ALPHA-D-MALTOSIDE, De novo designed serine hydrolase | | Authors: | Kuzin, A, Lew, S, Seetharaman, J, Maglaqui, M, Xiao, R, Kohan, E, Rajagopalan, S, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-05-08 | | Release date: | 2012-05-30 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.493 Å) | | Cite: | Design of activated serine-containing catalytic triads with atomic-level accuracy.
Nat.Chem.Biol., 10, 2014
|
|
6DDZ
 
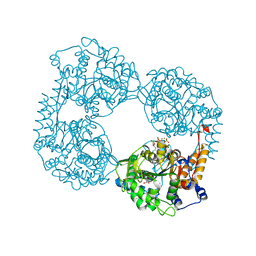 | | Crystal structure of the double mutant (D52N/R238W) of NT5C2-537X in the active state, Northeast Structural Genomics Target | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cytosolic purine 5'-nucleotidase, GLYCEROL, ... | | Authors: | Forouhar, F, Dieck, C.L, Tzoneva, G, Carpenter, Z, Ambesi-Impiombato, A, Sanchez-Martin, M, Kirschner-Schwabe, R, Lew, S, Seetharaman, J, Ferrando, A.A, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2018-05-10 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structure and Mechanisms of NT5C2 Mutations Driving Thiopurine Resistance in Relapsed Lymphoblastic Leukemia.
Cancer Cell, 34, 2018
|
|
3LMC
 
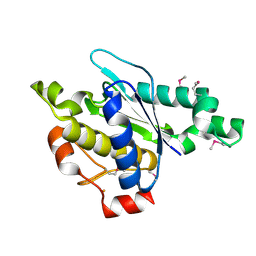 | | Crystal Structure of zinc-dependent peptidase from Methanocorpusculum labreanum (strain Z), Northeast Structural Genomics Consortium Target MuR16 | | Descriptor: | FE (III) ION, Peptidase, zinc-dependent, ... | | Authors: | Kuzin, A, Ashok, S, Vorobiev, S, Seetharaman, J, Patel, P, Xiao, R, Ciccosanti, C, Lee, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-01-29 | | Release date: | 2010-02-16 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Northeast Structural Genomics Consortium Target MuR16
To be published
|
|
1LMI
 
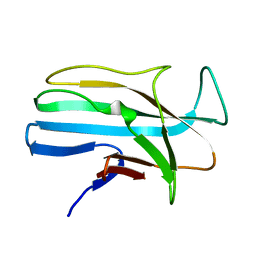 | | 1.5 ANGSTROM RESOLUTION CRYSTAL STRUCTURE OF A SECRETED PROTEIN FROM MYCOBACTERIUM TUBERCULOSIS-MPT63 | | Descriptor: | Immunogenic protein MPT63/MPB63 | | Authors: | Goulding, C.W, Parseghian, A, Sawaya, M.R, Cascio, D, Apostol, M, Gennaro, M.L, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2002-05-01 | | Release date: | 2002-12-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of a major secreted protein of Mycobacterium tuberculosis-MPT63 at
1.5-A resolution
Protein Sci., 11, 2002
|
|
3LJX
 
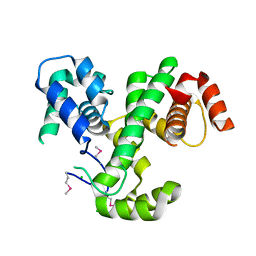 | | Crystal Structure of MmoQ Response regulator (fragment 20-298) from Methylococcus capsulatus str. Bath, Northeast Structural Genomics Consortium Target McR175G | | Descriptor: | CHLORIDE ION, MmoQ Response regulator | | Authors: | Kuzin, A, Scott, L, Forouhar, F, Abashidze, M, Seetharaman, J, Mao, M, Xiao, R, Ciccosanti, C, Wang, H, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-01-26 | | Release date: | 2010-02-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Northeast Structural Genomics Consortium Target McR175G
To be published, 2009
|
|
3LM6
 
 | | Crystal Structure of Stage V sporulation protein AD (spoVAD) from Bacillus subtilis, Northeast Structural Genomics Consortium Target SR525 | | Descriptor: | Stage V sporulation protein AD | | Authors: | Forouhar, F, Su, M, Seetharaman, J, Fang, F, Xiao, R, Cunningham, K, Ma, L, Wang, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-01-29 | | Release date: | 2010-02-16 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Northeast Structural Genomics Consortium Target SR525
To be Published
|
|
3LRQ
 
 | | Crystal structure of the U-box domain of human ubiquitin-protein ligase (E3), NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET HR4604D. | | Descriptor: | E3 ubiquitin-protein ligase TRIM37, ZINC ION | | Authors: | Kuzin, A, Chen, Y, Seetharaman, J, Mao, M, Xiao, R, Ciccosanti, C, Shastry, R, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-02-11 | | Release date: | 2010-03-23 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.292 Å) | | Cite: | E3 UBIQUITIN-PROTEIN LIGASE RING DOMAIN FROM HUMAN TRIPARTITE MOTIF-CONTAINING PROTEIN 37 (TRIM37), NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET HR4604D.
To be Published
|
|
2RJE
 
 | | Crystal structure of L3MBTL1 in complex with H4K20Me2 (residues 17-25), orthorhombic form II | | Descriptor: | CHLORIDE ION, Histone H4, Lethal(3)malignant brain tumor-like protein | | Authors: | Allali-Hassani, A, Liu, Y, Herzanych, N, Ouyang, H, Mackenzie, F, Crombet, L, Loppnau, P, Kozieradzki, I, Vedadi, M, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J.R, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-10-14 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | L3MBTL1 recognition of mono- and dimethylated histones.
Nat.Struct.Mol.Biol., 14, 2007
|
|
6SHZ
 
 | | p53 cancer mutant Y220C | | Descriptor: | 1,2-ETHANEDIOL, Cellular tumor antigen p53, GLYCEROL, ... | | Authors: | Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-08-08 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Targeting Cavity-Creating p53 Cancer Mutations with Small-Molecule Stabilizers: the Y220X Paradigm.
Acs Chem.Biol., 15, 2020
|
|
4XY9
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a 2-amine-9H-purine ligand | | Descriptor: | 1,2-ETHANEDIOL, 6-(5-bromo-2-methoxyphenyl)-9H-purin-2-amine, Bromodomain-containing protein 4 | | Authors: | Picaud, S, von Delft, F, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Filippakopoulos, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-02-02 | | Release date: | 2015-03-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | 9H-Purine Scaffold Reveals Induced-Fit Pocket Plasticity of the BRD9 Bromodomain.
J.Med.Chem., 58, 2015
|
|
1P3H
 
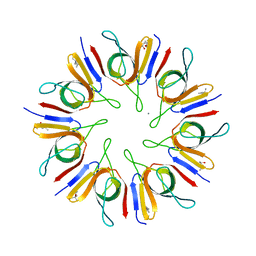 | | Crystal Structure of the Mycobacterium tuberculosis chaperonin 10 tetradecamer | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 10 kDa chaperonin, CALCIUM ION | | Authors: | Roberts, M.M, Coker, A.R, Fossati, G, Mascagni, P, Coates, A.R.M, Wood, S.P, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2003-04-17 | | Release date: | 2003-07-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mycobacterium tuberculosis chaperonin 10 heptamers self-associate through their biologically active loops
J.BACTERIOL., 185, 2003
|
|
3KLU
 
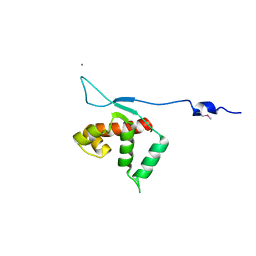 | | Crystal structure of the protein yqbn. northeast structural genomics consortium target sr445. | | Descriptor: | MANGANESE (II) ION, Uncharacterized protein yqbN | | Authors: | Kuzin, A.P, Su, M, Seetharaman, J, Wang, D, Fang, Y, Cunningham, K, Ma, L.-C, Xiao, R, Liu, M, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-11-09 | | Release date: | 2009-11-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the protein yqbn. northeast structural genomics consortium target sr445.
To be Published
|
|
4XYA
 
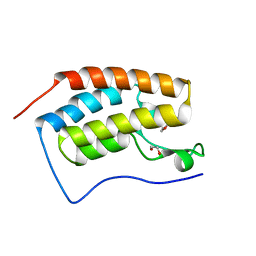 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a 2-amine-9H-purine ligand | | Descriptor: | 1,2-ETHANEDIOL, 6-(5-bromo-1-benzofuran-7-yl)-9H-purin-2-amine, Bromodomain-containing protein 4 | | Authors: | Picaud, S, von Delft, F, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Filippakopoulos, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-02-02 | | Release date: | 2015-03-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | 9H-Purine Scaffold Reveals Induced-Fit Pocket Plasticity of the BRD9 Bromodomain.
J.Med.Chem., 58, 2015
|
|
4XY8
 
 | | Crystal Structure of the bromodomain of BRD9 in complex with a 2-amine-9H-purine ligand | | Descriptor: | 6-(5-bromo-2-methoxyphenyl)-9H-purin-2-amine, BROMIDE ION, Bromodomain-containing protein 9 | | Authors: | Picaud, S, von Delft, F, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Filippakopoulos, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-02-02 | | Release date: | 2015-03-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 9H-Purine Scaffold Reveals Induced-Fit Pocket Plasticity of the BRD9 Bromodomain.
J.Med.Chem., 58, 2015
|
|
3K2T
 
 | | Crystal structure of Lmo2511 protein from Listeria monocytogenes, northeast structural genomics consortium target LkR84A | | Descriptor: | Lmo2511 protein | | Authors: | Seetharaman, J, Su, M, Wang, D, Janjua, H, Cunningham, K, Owens, L, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-09-30 | | Release date: | 2009-11-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of Lmo2511 protein from Listeria monocytogenes, northeast structural genomics consortium target LkR84A
To be Published
|
|
6BU1
 
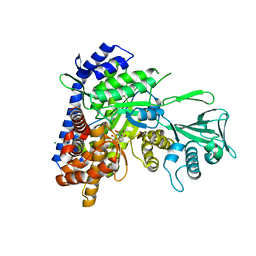 | |
4MPS
 
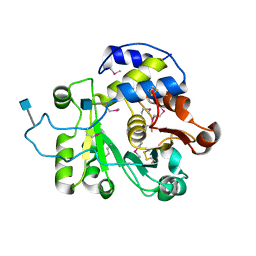 | | Crystal structure of rat Beta-galactoside alpha-2,6-sialyltransferase 1 (ST6GAL1), Northeast Structural Genomics Consortium Target RnR367A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-galactoside alpha-2,6-sialyltransferase 1 | | Authors: | Forouhar, F, Meng, L, Milaninia, S, Seetharaman, J, Su, M, Kornhaber, G, Montelione, G.T, Hunt, J.F, Moremen, K.W, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-09-13 | | Release date: | 2013-09-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Enzymatic Basis for N-Glycan Sialylation: STRUCTURE OF RAT alpha 2,6-SIALYLTRANSFERASE (ST6GAL1) REVEALS CONSERVED AND UNIQUE FEATURES FOR GLYCAN SIALYLATION.
J.Biol.Chem., 288, 2013
|
|
3I3U
 
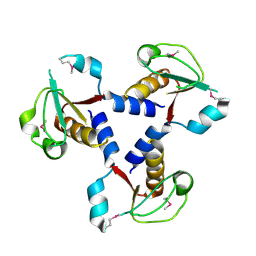 | | Crystal structure of lp_1913 protein from lactobacillus plantarum, northeast structural genomics Consortium target lpr140a | | Descriptor: | uncharacterized protein lp_1913 | | Authors: | Seetharaman, J, Chen, Y, Forouhar, F, Sahdev, S, Janjua, H, Xiao, R, Ciccosanti, C, Foote, E.L, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-06-30 | | Release date: | 2009-07-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of lp_1913 protein from lactobacillus plantarum, northeast structural genomics Consortium target lpr140a
To be Published
|
|
2AKL
 
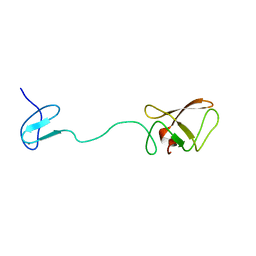 | | Solution structure for phn-A like protein PA0128 from Pseudomonas aeruginosa | | Descriptor: | ZINC ION, phnA-like protein pa0128 | | Authors: | Srisailam, S, Yee, A, Lemak, A, Lukin, J.A, Bansal, S, Prestegard, J.H, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG), Ontario Centre for Structural Proteomics (OCSP) | | Deposit date: | 2005-08-03 | | Release date: | 2006-07-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Sequence Specific Resonance Assignment of a Hypothetical Protein PA0128 from Pseudomonas Aeruginosa
J.Biomol.Nmr, 36, 2006
|
|
6X9O
 
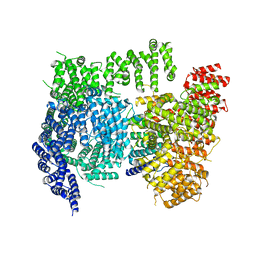 | | High resolution cryoEM structure of huntingtin in complex with HAP40 | | Descriptor: | 40-kDa huntingtin-associated protein, Huntingtin | | Authors: | Harding, R.J, Deme, J.C, Lea, S.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-06-03 | | Release date: | 2020-06-17 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Huntingtin structure is orchestrated by HAP40 and shows a polyglutamine expansion-specific interaction with exon 1.
Commun Biol, 4, 2021
|
|
1OY0
 
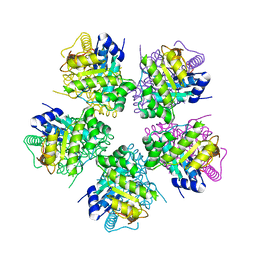 | | The crystal Structure of the First Enzyme of Pantothenate Biosynthetic Pathway, Ketopantoate Hydroxymethyltransferase from Mycobacterium Tuberculosis Shows a Decameric Assembly and Terminal Helix-Swapping | | Descriptor: | Ketopantoate hydroxymethyltransferase, MAGNESIUM ION | | Authors: | Chaudhuri, B.N, Sawaya, M.R, Kim, C.Y, Waldo, G.S, Park, M.S, Terwilliger, T.C, Yeates, T.O, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2003-04-03 | | Release date: | 2003-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Crystal Structure of the First Enzyme in the Pantothenate Biosynthetic Pathway,
Ketopantoate Hydroxymethyltransferase, from M. tuberculosis
Structure, 11, 2003
|
|
3L6I
 
 | | Crystal structure of the uncharacterized lipoprotein yceb from e. coli at the resolution 2.0a. northeast structural genomics consortium target er542 | | Descriptor: | SODIUM ION, Uncharacterized lipoprotein yceB | | Authors: | Kuzin, A.P, Neely, H, Seetharaman, J, Chen, C.X, Janjua, H, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-12-23 | | Release date: | 2010-01-26 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.011 Å) | | Cite: | Crystal structure of the uncharacterized lipoprotein yceb from e. coli at the resolution 2.0a. northeast structural genomics consortium target er542
To be Published
|
|
