5DEM
 
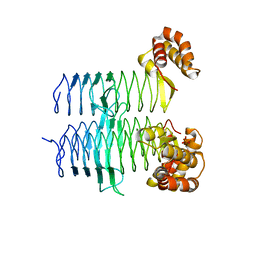 | | Structure of Pseudomonas aeruginosa LpxA | | Descriptor: | Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, PHOSPHATE ION | | Authors: | Smith, E.W, Chen, Y. | | Deposit date: | 2015-08-25 | | Release date: | 2015-09-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structures of Pseudomonas aeruginosa LpxA Reveal the Basis for Its Substrate Selectivity.
Biochemistry, 54, 2015
|
|
8P5Y
 
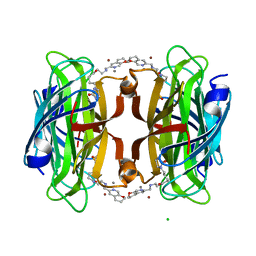 | | Artificial transfer hydrogenase with a Mn-12 cofactor and Streptavidin S112Y-K121M mutant | | Descriptor: | 5-[(3~{a}~{S},4~{S},6~{a}~{R})-2-oxidanylidene-1,3,3~{a},4,6,6~{a}-hexahydrothieno[3,4-d]imidazol-4-yl]-~{N}-[2-(3,4-dihydro-2~{H}-pyrano[2,3-c]pyridin-6-ylmethylamino)ethyl]pentanamide, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Lau, K, Wang, W, Pojer, F, Larabi, A. | | Deposit date: | 2023-05-24 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Manganese Transfer Hydrogenases Based on the Biotin-Streptavidin Technology.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8P5Z
 
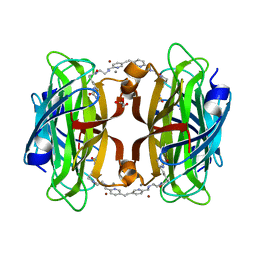 | | Artificial transfer hydrogenase with a Mn-5 cofactor and Streptavidin S112Y-K121M mutant | | Descriptor: | 5-[(3~{a}~{S},4~{S},6~{a}~{R})-2-oxidanylidene-1,3,3~{a},4,6,6~{a}-hexahydrothieno[3,4-d]imidazol-4-yl]-~{N}-[2-[(5-methylpyridin-2-yl)methylamino]ethyl]pentanamide, BROMIDE ION, GLYCEROL, ... | | Authors: | Lau, K, Wang, W, Pojer, F, Larabi, A. | | Deposit date: | 2023-05-24 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Manganese Transfer Hydrogenases Based on the Biotin-Streptavidin Technology.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7ESI
 
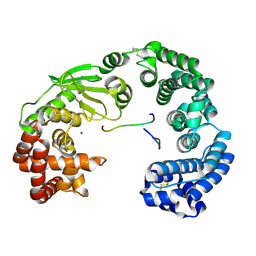 | | Crystal structure of the collagenase unit of a Vibrio collagenase from Vibrio harveyi VHJR7 at 1. 8 angstrom resolution. | | Descriptor: | CALCIUM ION, Collagenase unit (CU), Peptide P1, ... | | Authors: | Cao, H.Y, Wang, Y, Peng, M, Zhang, Y.Z. | | Deposit date: | 2021-05-11 | | Release date: | 2022-02-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Vibrio collagenase VhaC provides insight into the mechanism of bacterial collagenolysis.
Nat Commun, 13, 2022
|
|
3HDB
 
 | | Crystal structure of AaHIV, A metalloproteinase from venom of Agkistrodon Acutus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, AAHIV, CALCIUM ION, ... | | Authors: | Zhu, Z.Q, Niu, L.W, Teng, M.K. | | Deposit date: | 2009-05-07 | | Release date: | 2009-08-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural basis of the autolysis of AaHIV suggests a novel target recognizing model for ADAM/reprolysin family proteins
Biochem.Biophys.Res.Commun., 386, 2009
|
|
5DZO
 
 | |
8QPR
 
 | | SARS-CoV-2 S protein bound to human neutralising antibody UZGENT_G5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, IgG heavy chain - FAB, ... | | Authors: | Remaut, H, Reiter, D, Vandenkerckhove, L, Acar, D.D, Witkowski, W, Gerlo, S. | | Deposit date: | 2023-10-03 | | Release date: | 2024-01-31 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Integrating artificial intelligence-based epitope prediction in a SARS-CoV-2 antibody discovery pipeline: caution is warranted.
Ebiomedicine, 100, 2024
|
|
8QQ0
 
 | | SARS-CoV-2 S protein bound to neutralising antibody UZGENT_A3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Remaut, H, Reiter, D, Vandenkerckhove, L, Acar, D.D, Witkowski, W, Gerlo, S. | | Deposit date: | 2023-10-03 | | Release date: | 2024-01-31 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Integrating artificial intelligence-based epitope prediction in a SARS-CoV-2 antibody discovery pipeline: caution is warranted.
Ebiomedicine, 100, 2024
|
|
1AKN
 
 | | STRUCTURE OF BILE-SALT ACTIVATED LIPASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BILE-SALT ACTIVATED LIPASE | | Authors: | Wang, X, Zhang, X. | | Deposit date: | 1997-05-23 | | Release date: | 1998-05-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of bovine bile salt activated lipase: insights into the bile salt activation mechanism.
Structure, 5, 1997
|
|
1AQL
 
 | |
4N5G
 
 | | Crystal Structure of RXRa LBD complexed with a synthetic modulator K8012 | | Descriptor: | 5-(2-{(1Z)-5-fluoro-2-methyl-1-[4-(propan-2-yl)benzylidene]-1H-inden-3-yl}ethyl)-1H-tetrazole, Retinoic acid receptor RXR-alpha | | Authors: | Aleshin, A.E, Su, Y, Zhang, X, Liddington, R.C. | | Deposit date: | 2013-10-09 | | Release date: | 2014-05-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Sulindac-Derived RXR alpha Modulators Inhibit Cancer Cell Growth by Binding to a Novel Site.
Chem.Biol., 21, 2014
|
|
5DZN
 
 | | human T-cell immunoglobulin and mucin domain protein 4 | | Descriptor: | T-cell immunoglobulin and mucin domain-containing protein 4 | | Authors: | Yuan, S, Rao, Z, Wang, X. | | Deposit date: | 2015-09-25 | | Release date: | 2015-11-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | TIM-1 acts a dual-attachment receptor for Ebolavirus by interacting directly with viral GP and the PS on the viral envelope.
Protein Cell, 6, 2015
|
|
3GZT
 
 | | VP7 recoated rotavirus DLP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Outer capsid glycoprotein VP7 | | Authors: | Chen, J.Z, Settembre, E.C, Harrison, S.C, Grigorieff, N. | | Deposit date: | 2009-04-07 | | Release date: | 2009-07-14 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Molecular interactions in rotavirus assembly and uncoating seen by high-resolution cryo-EM
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3GZU
 
 | | VP7 recoated rotavirus DLP | | Descriptor: | Inner capsid protein VP2, Intermediate capsid protein VP6, ZINC ION | | Authors: | Chen, J.Z, Settembre, E.C, Harrison, S.C, Grigorieff, N. | | Deposit date: | 2009-04-07 | | Release date: | 2009-07-14 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Molecular interactions in rotavirus assembly and uncoating seen by high-resolution cryo-EM.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
8UAB
 
 | | SARS-CoV-2 main protease (Mpro) complex with AC1115 | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-1-({(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-1H-indole-2-carboxamide | | Authors: | DuPrez, K.T, Chao, A, Han, F.Q. | | Deposit date: | 2023-09-20 | | Release date: | 2024-02-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.781 Å) | | Cite: | Olgotrelvir, a dual inhibitor of SARS-CoV-2 M pro and cathepsin L, as a standalone antiviral oral intervention candidate for COVID-19
Med, 5, 2024
|
|
8UAC
 
 | | CATHEPSIN L IN COMPLEX WITH AC1115 | | Descriptor: | Cathepsin L, N-[(2S)-1-({(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-1H-indole-2-carboxamide | | Authors: | Chao, A, DuPrez, K.T, Han, F.Q. | | Deposit date: | 2023-09-20 | | Release date: | 2024-02-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Olgotrelvir, a dual inhibitor of SARS-CoV-2 M pro and cathepsin L, as a standalone antiviral oral intervention candidate for COVID-19.
Med, 5, 2024
|
|
3NBV
 
 | |
3JUA
 
 | |
4N8R
 
 | | Crystal structure of RXRa LBD complexed with a synthetic modulator K-8008 | | Descriptor: | 5-(2-{(1Z)-2-methyl-1-[4-(propan-2-yl)benzylidene]-1H-inden-3-yl}ethyl)-1H-tetrazole, Retinoic acid receptor RXR-alpha | | Authors: | Aleshin, A.E, Su, Y, Zhang, X, Liddington, R.C. | | Deposit date: | 2013-10-17 | | Release date: | 2014-05-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Sulindac-Derived RXR alpha Modulators Inhibit Cancer Cell Growth by Binding to a Novel Site.
Chem.Biol., 21, 2014
|
|
3NBW
 
 | | X-ray structure of ketohexokinase in complex with a pyrazole compound | | Descriptor: | 5-amino-3-(methylsulfanyl)-1-phenyl-1H-pyrazole-4-carbonitrile, GLYCEROL, Ketohexokinase, ... | | Authors: | Abad, M.C, Gibbs, A.C, Spurlino, J.C. | | Deposit date: | 2010-06-04 | | Release date: | 2010-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.341 Å) | | Cite: | Electron density guided fragment-based lead discovery of ketohexokinase inhibitors.
J.Med.Chem., 53, 2010
|
|
3NC2
 
 | |
3NC9
 
 | |
3J2Z
 
 | |
3NMU
 
 | | Crystal Structure of substrate-bound halfmer box C/D RNP | | Descriptor: | 50S ribosomal protein L7Ae, Fibrillarin-like rRNA/tRNA 2'-O-methyltransferase, NOP5/NOP56 related protein, ... | | Authors: | Li, H, Xue, S, Wang, R. | | Deposit date: | 2010-06-22 | | Release date: | 2011-05-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.729 Å) | | Cite: | Structural basis for substrate placement by an archaeal box C/D ribonucleoprotein particle.
Mol.Cell, 39, 2010
|
|
3NCA
 
 | | X-ray structure of ketohexokinase in complex with a thieno pyridinol compound | | Descriptor: | Ketohexokinase, SULFATE ION, thieno[3,2-b]pyridin-7-ol | | Authors: | Abad, M.C, Gibbs, A.C, Spurlino, J.C. | | Deposit date: | 2010-06-04 | | Release date: | 2010-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.602 Å) | | Cite: | Electron density guided fragment-based lead discovery of ketohexokinase inhibitors.
J.Med.Chem., 53, 2010
|
|
