8WM4
 
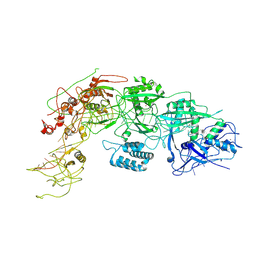 | | Cryo-EM structure of DiCas7-11 in complex with crRNA | | Descriptor: | CRISPR-associated RAMP family protein, ZINC ION, crRNA (38-MER) | | Authors: | Ma, H.Y, Tang, X.D. | | Deposit date: | 2023-10-02 | | Release date: | 2024-05-29 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structural basis of negative regulation of CRISPR-Cas7-11 by TPR-CHAT.
Signal Transduct Target Ther, 9, 2024
|
|
8WML
 
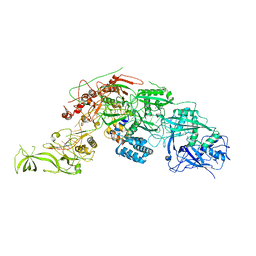 | |
8WMC
 
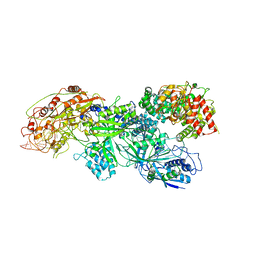 | | Cryo-EM structure of DiCas7-11-crRNA in complex with regulator | | Descriptor: | CHAT domain-containing protein, CRISPR-associated RAMP family protein, RNA (38-MER), ... | | Authors: | Ma, H.Y, Tang, X.D. | | Deposit date: | 2023-10-03 | | Release date: | 2024-05-29 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of negative regulation of CRISPR-Cas7-11 by TPR-CHAT.
Signal Transduct Target Ther, 9, 2024
|
|
8XKL
 
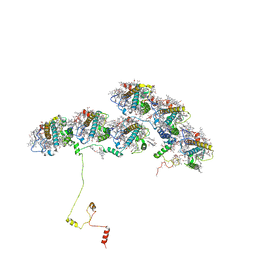 | | Structure of ACPII-CCPII from cryptophyte algae | | Descriptor: | (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E})-3,7,12,16-tetramethyl-18-[(4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-3,5,7,9,11,13,15-heptaen-1,17-diynyl]cyclohex-3-en-1-ol, (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-(2,6,6-trimethylcyclohexen-1-yl)octadeca-3,5,7,9,11,13,15,17-octaen-1-ynyl]cyclohex-3-en-1-ol, (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-[(1~{R},4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohex-2-en-1-yl]octadeca-3,5,7,9,11,13,15,17-octaen-1-ynyl]cyclohex-3-en-1-ol, ... | | Authors: | Li, X.Y, Mao, Z.Y, Shen, J.R, Han, G.Y. | | Deposit date: | 2023-12-23 | | Release date: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Structure and distinct supramolecular organization of a PSII-ACPII dimer from a cryptophyte alga Chroomonas placoidea.
Nat Commun, 15, 2024
|
|
5TSR
 
 | |
6UF7
 
 | | S2 symmetric peptide design number 5, Uncle Fester | | Descriptor: | S2-5, Uncle Fester | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-23 | | Release date: | 2020-12-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (0.8 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
6UF8
 
 | | S2 symmetric peptide design number 6, London Bridge | | Descriptor: | S2-6, London Bridge | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-23 | | Release date: | 2020-12-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (0.8 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
6UFU
 
 | | C2 symmetric peptide design number 1, Zappy, crystal form 1 | | Descriptor: | C2-1, Zappy, crystal form 1 | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-25 | | Release date: | 2020-12-02 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.101 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
6UG3
 
 | | C3 symmetric peptide design number 1, Sporty, crystal form 1 | | Descriptor: | C3-1, Sporty, crystal form 1, ... | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-25 | | Release date: | 2020-12-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
6UGC
 
 | | C3 symmetric peptide design number 3 | | Descriptor: | C3-3 cyclic peptide design, CADMIUM ION, SODIUM ION | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-26 | | Release date: | 2020-12-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
6UDR
 
 | | S2 symmetric peptide design number 3 crystal form 1, Lurch | | Descriptor: | S2-3, Lurch crystal form 1 | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-19 | | Release date: | 2020-09-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
6UD9
 
 | | S2 symmetric peptide design number 2, Morticia | | Descriptor: | S2-2, Morticia | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-19 | | Release date: | 2020-09-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
6UCX
 
 | | S2 symmetric peptide design number 1, Wednesday | | Descriptor: | S2-1, Wednesday, trifluoroacetic acid | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-18 | | Release date: | 2020-09-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
6UDZ
 
 | | S2 symmetric peptide design number 4 crystal form 1, Pugsley | | Descriptor: | S2-4, Pusgley crystal form 1, trifluoroacetic acid | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-20 | | Release date: | 2020-09-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
5UY1
 
 | | X-ray crystal structure of apo Halotag | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Haloalkane dehalogenase | | Authors: | Dunham, N.P, Boal, A.K. | | Deposit date: | 2017-02-23 | | Release date: | 2017-03-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The Cation-pi Interaction Enables a Halo-Tag Fluorogenic Probe for Fast No-Wash Live Cell Imaging and Gel-Free Protein Quantification.
Biochemistry, 56, 2017
|
|
5UXZ
 
 | |
5UN6
 
 | |
2F2O
 
 | |
5UN5
 
 | |
7XWX
 
 | | Crystal structure of SARS-CoV-2 N-CTD | | Descriptor: | Nucleoprotein, PHOSPHATE ION | | Authors: | Luan, X.D, Li, X.M, Li, Y.F. | | Deposit date: | 2022-05-27 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Antiviral drug design based on structural insights into the N-terminal domain and C-terminal domain of the SARS-CoV-2 nucleocapsid protein.
Sci Bull (Beijing), 67, 2022
|
|
5V45
 
 | |
5V46
 
 | | Crystal structure of the I113M, F270M, K291M, L308M mutant of SR1 domain of human sacsin | | Descriptor: | Sacsin | | Authors: | Menade, M, Kozlov, G, Gehring, K. | | Deposit date: | 2017-03-08 | | Release date: | 2017-05-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of ubiquitin-like (Ubl) and Hsp90-like domains of sacsin provide insight into pathological mutations.
J. Biol. Chem., 293, 2018
|
|
2F2P
 
 | |
1WP8
 
 | | crystal structure of Hendra Virus fusion core | | Descriptor: | Fusion glycoprotein F0,Fusion glycoprotein F0 | | Authors: | Xu, Y, Liu, Y, Lou, Z, Su, N, Bai, Z, Gao, G.F, Rao, Z. | | Deposit date: | 2004-08-31 | | Release date: | 2005-11-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of Nipah and Hendra virus fusion core proteins
FEBS J., 273, 2006
|
|
6KI7
 
 | | Pyrophosphatase mutant K30R from Acinetobacter baumannii | | Descriptor: | Inorganic pyrophosphatase | | Authors: | Su, J. | | Deposit date: | 2019-07-17 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal Structures of Pyrophosphatase from Acinetobacter baumannii: Snapshots of Pyrophosphate Binding and Identification of a Phosphorylated Enzyme Intermediate.
Int J Mol Sci, 20, 2019
|
|
