4DJW
 
 | |
4DJU
 
 | | Structure of BACE Bound to 2-imino-3-methyl-5,5-diphenylimidazolidin-4-one | | Descriptor: | (2E)-2-imino-3-methyl-5,5-diphenylimidazolidin-4-one, Beta-secretase 1, L(+)-TARTARIC ACID | | Authors: | Strickland, C, Cumming, J. | | Deposit date: | 2012-02-02 | | Release date: | 2012-03-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure based design of iminohydantoin BACE1 inhibitors: Identification of an orally available, centrally active BACE1 inhibitor.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4DJX
 
 | |
4FRS
 
 | |
4DJY
 
 | |
4DJV
 
 | | Structure of BACE Bound to 2-imino-5-(3'-methoxy-[1,1'-biphenyl]-3-yl)-3-methyl-5-phenylimidazolidin-4-one | | Descriptor: | (2E,5R)-2-imino-5-(3'-methoxybiphenyl-3-yl)-3-methyl-5-phenylimidazolidin-4-one, Beta-secretase 1, L(+)-TARTARIC ACID | | Authors: | Strickland, C, Cumming, J. | | Deposit date: | 2012-02-02 | | Release date: | 2012-03-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structure based design of iminohydantoin BACE1 inhibitors: Identification of an orally available, centrally active BACE1 inhibitor.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4L53
 
 | | Crystal Structure of (1R,4R)-4-{4-[7-amino-2-(1,2,3-benzothiadiazol-7-yl)-3-chlorofuro[2,3-c]pyridin-4-yl]-1H-pyrazol-1-yl}cyclohexan-1-ol bound to TAK1-TAB1 | | Descriptor: | 1,2-ETHANEDIOL, Mitogen-activated protein kinase kinase kinase 7, TGF-beta-activated kinase 1 and MAP3K7-binding protein 1 chimera, ... | | Authors: | Wang, J, Hornberger, K.R, Crew, A.P, Jestel, A, Maskos, K, Moertl, M. | | Deposit date: | 2013-06-10 | | Release date: | 2013-07-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Discovery of 7-aminofuro[2,3-c]pyridine inhibitors of TAK1: Optimization of kinase selectivity and pharmacokinetics.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
5C1U
 
 | | Crystal structure of EV71 3C Proteinase in complex with Compound Xb | | Descriptor: | (2S)-2-[[(E)-3-[4-(dimethylamino)phenyl]prop-2-enoyl]amino]-N-[(2S)-1-oxidanyl-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]-3-phenyl-propanamide, 3C proteinase | | Authors: | Zhang, L, Huang, G, Cai, Q, Zhao, C, Ren, H, Li, P, Li, N, Chen, S, Li, J, Lin, T. | | Deposit date: | 2015-06-15 | | Release date: | 2016-06-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Optimize the interactions at S4 with efficient inhibitors targeting 3C proteinase from enterovirus 71
J.Mol.Recognit., 29, 2016
|
|
5C1X
 
 | | Crystal structure of EV71 3C Proteinase in complex with Compound VIII | | Descriptor: | (phenylmethyl) N-[(2S)-1-oxidanylidene-1-[[(2S)-1-oxidanyl-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-3-phenyl-propan-2-yl]carbamate, 3C proteinase | | Authors: | Zhang, L, Huang, G, Cai, Q, Zhao, C, Ren, H, Li, P, Li, N, Chen, S, Li, J, Lin, T. | | Deposit date: | 2015-06-15 | | Release date: | 2016-06-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Optimize the interactions at S4 with efficient inhibitors targeting 3C proteinase from enterovirus 71
J.Mol.Recognit., 29, 2016
|
|
5C8Y
 
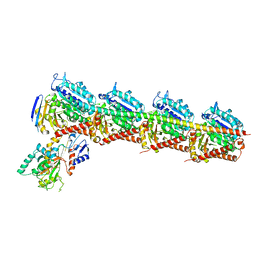 | | Crystal structure of T2R-TTL-Plinabulin complex | | Descriptor: | (3Z,6Z)-3-benzylidene-6-[(5-tert-butyl-1H-imidazol-4-yl)methylidene]piperazine-2,5-dione, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wang, Y, Yu, Y, Chen, Q, Yang, J. | | Deposit date: | 2015-06-26 | | Release date: | 2015-11-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.594 Å) | | Cite: | Structures of a diverse set of colchicine binding site inhibitors in complex with tubulin provide a rationale for drug discovery.
Febs J., 283, 2016
|
|
8HC5
 
 | | SARS-CoV-2 wildtype S1 in complex with YB9-258 Fab and R1-32 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R1-32 Fab, Heavy chain of YB9-258 Fab, ... | | Authors: | Liu, B, Gao, X, Chen, Q, Li, Z, Su, M, He, J, Xiong, X. | | Deposit date: | 2022-11-01 | | Release date: | 2023-01-25 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | Somatically hypermutated antibodies isolated from SARS-CoV-2 Delta infected patients cross-neutralize heterologous variants.
Nat Commun, 14, 2023
|
|
8HCB
 
 | | SARS-CoV-2 Omicron BA.1 spike trimer (6P) in complex with 3 YB13-292 Fabs (2 RBD up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of YB13-292 Fab, ... | | Authors: | Liu, B, Gao, X, Chen, Q, Li, Z, Su, M, He, J, Xiong, X. | | Deposit date: | 2022-11-01 | | Release date: | 2023-01-25 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.18 Å) | | Cite: | Somatically hypermutated antibodies isolated from SARS-CoV-2 Delta infected patients cross-neutralize heterologous variants.
Nat Commun, 14, 2023
|
|
8HC8
 
 | | SARS-CoV-2 Omicron BA.1 spike trimer (6P) in complex with YB13-292 Fab, focused refinement of Fab region | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of YB13-292 Fab, Light chain of YB13-292 Fab, ... | | Authors: | Liu, B, Gao, X, Chen, Q, Li, Z, Su, M, He, J, Xiong, X. | | Deposit date: | 2022-11-01 | | Release date: | 2023-01-25 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.95 Å) | | Cite: | Somatically hypermutated antibodies isolated from SARS-CoV-2 Delta infected patients cross-neutralize heterologous variants.
Nat Commun, 14, 2023
|
|
8HC7
 
 | | SARS-CoV-2 Omicron BA.1 spike trimer (6P) complex with YB9-258 Fab, focused refinement of RBD-dimer region | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain variable region of YB9-258, Light chain variable region of YB9-258, ... | | Authors: | Liu, B, Gao, X, Chen, Q, Li, Z, Su, M, He, J, Xiong, X. | | Deposit date: | 2022-11-01 | | Release date: | 2023-01-25 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.54 Å) | | Cite: | Somatically hypermutated antibodies isolated from SARS-CoV-2 Delta infected patients cross-neutralize heterologous variants.
Nat Commun, 14, 2023
|
|
8HC3
 
 | | SARS-CoV-2 Omicron BA.1 spike trimer (6P) in complex with 2 YB9-258 Fabs (2 RBD up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of YB9-258, ... | | Authors: | Liu, B, Gao, X, Chen, Q, Li, Z, Su, M, He, J, Xiong, X. | | Deposit date: | 2022-11-01 | | Release date: | 2023-01-25 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.35 Å) | | Cite: | Somatically hypermutated antibodies isolated from SARS-CoV-2 Delta infected patients cross-neutralize heterologous variants.
Nat Commun, 14, 2023
|
|
8HC4
 
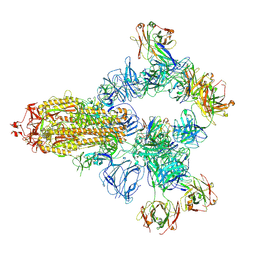 | | SARS-CoV-2 wildtype spike trimer (6P) in complex with 3 YB9-258 Fabs and 3 R1-32 Fabs (3 RBD up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R1-32 Fab, ... | | Authors: | Liu, B, Gao, X, Chen, Q, Li, Z, Su, M, He, J, Xiong, X. | | Deposit date: | 2022-11-01 | | Release date: | 2023-01-25 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Somatically hypermutated antibodies isolated from SARS-CoV-2 Delta infected patients cross-neutralize heterologous variants.
Nat Commun, 14, 2023
|
|
8HCA
 
 | | SARS-CoV-2 Omicron BA.1 spike trimer (6P) in complex with 3 YB13-292 Fabs (1 RBD up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of YB13-292 Fab, ... | | Authors: | Liu, B, Gao, X, Chen, Q, Li, Z, Su, M, He, J, Xiong, X. | | Deposit date: | 2022-11-01 | | Release date: | 2023-01-25 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (4.35 Å) | | Cite: | Somatically hypermutated antibodies isolated from SARS-CoV-2 Delta infected patients cross-neutralize heterologous variants.
Nat Commun, 14, 2023
|
|
5CA1
 
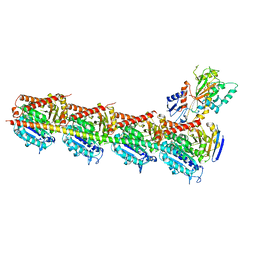 | | Crystal structure of T2R-TTL-Nocodazole complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Wang, Y, Yu, Y, Chen, Q, Yang, J. | | Deposit date: | 2015-06-29 | | Release date: | 2015-11-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Structures of a diverse set of colchicine binding site inhibitors in complex with tubulin provide a rationale for drug discovery.
Febs J., 283, 2016
|
|
8HC6
 
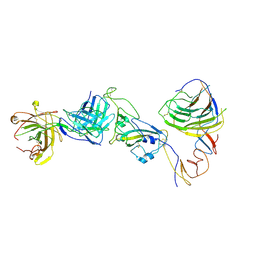 | | SARS-CoV-2 Omicron BA.1 spike trimer (6P) in complex with YB9-258 Fab, focused refinement of Fab region | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of YB9-258 Fab, Light chain of YB9-258, ... | | Authors: | Liu, B, Gao, X, Chen, Q, Li, Z, Su, M, He, J, Xiong, X. | | Deposit date: | 2022-11-01 | | Release date: | 2023-01-25 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.69 Å) | | Cite: | Somatically hypermutated antibodies isolated from SARS-CoV-2 Delta infected patients cross-neutralize heterologous variants.
Nat Commun, 14, 2023
|
|
5CA0
 
 | | Crystal structure of T2R-TTL-Lexibulin complex | | Descriptor: | 1-ethyl-3-[2-methoxy-4-(5-methyl-4-{[(1S)-1-(pyridin-3-yl)butyl]amino}pyrimidin-2-yl)phenyl]urea, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wang, Y, Yu, Y, Chen, Q, Yang, J. | | Deposit date: | 2015-06-29 | | Release date: | 2015-11-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Structures of a diverse set of colchicine binding site inhibitors in complex with tubulin provide a rationale for drug discovery.
Febs J., 283, 2016
|
|
6IR2
 
 | | Crystal structure of red fluorescent protein mCherry complexed with the nanobody LaM2 at 1.4 Angstron resolution | | Descriptor: | MCherry fluorescent protein, mCherry's nanobody LaM2 | | Authors: | Ding, Y, Wang, Z.Y, Hu, R.T, Chen, X. | | Deposit date: | 2018-11-09 | | Release date: | 2019-11-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.393 Å) | | Cite: | Structural insights into the binding of nanobodies LaM2 and LaM4 to the red fluorescent protein mCherry.
Protein Sci., 30, 2021
|
|
5CB4
 
 | | Crystal structure of T2R-TTL-Tivantinib complex | | Descriptor: | (3R,4R)-3-(5,6-dihydro-4H-pyrrolo[3,2,1-ij]quinolin-1-yl)-4-(1H-indol-3-yl)pyrrolidine-2,5-dione, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wang, Y, Yu, Y, Chen, Q, Yang, J. | | Deposit date: | 2015-06-30 | | Release date: | 2015-11-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.193 Å) | | Cite: | Structures of a diverse set of colchicine binding site inhibitors in complex with tubulin provide a rationale for drug discovery.
Febs J., 283, 2016
|
|
8HC9
 
 | | SARS-CoV-2 Omicron BA.1 spike trimer (6P) in complex with 3 YB13-292 Fabs (3 RBD down) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of YB13-292 Fab, ... | | Authors: | Liu, B, Gao, X, Chen, Q, Li, Z, Su, M, He, J, Xiong, X. | | Deposit date: | 2022-11-01 | | Release date: | 2023-01-25 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (6.03 Å) | | Cite: | Somatically hypermutated antibodies isolated from SARS-CoV-2 Delta infected patients cross-neutralize heterologous variants.
Nat Commun, 14, 2023
|
|
8HC2
 
 | | SARS-CoV-2 Omicron BA.1 spike trimer (6P) in complex with 1 YB9-258 Fab (1 RBD up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of YB9-258 Fab, ... | | Authors: | Liu, B, Gao, X, Chen, Q, Li, Z, Su, M, He, J, Xiong, X. | | Deposit date: | 2022-11-01 | | Release date: | 2023-01-25 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (6.21 Å) | | Cite: | Somatically hypermutated antibodies isolated from SARS-CoV-2 Delta infected patients cross-neutralize heterologous variants.
Nat Commun, 14, 2023
|
|
6IR1
 
 | | Crystal structure of red fluorescent protein mCherry complexed with the nanobody LaM4 at 1.9 Angstron resolution | | Descriptor: | MCherry fluorescent protein, mCherry's nanobody LaM4 | | Authors: | Ding, Y, Wang, Z.Y, Hu, R.T, Chen, X. | | Deposit date: | 2018-11-09 | | Release date: | 2019-11-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.919 Å) | | Cite: | Structural insights into the binding of nanobodies LaM2 and LaM4 to the red fluorescent protein mCherry.
Protein Sci., 30, 2021
|
|
