6GYO
 
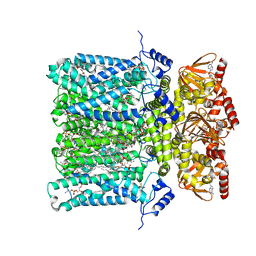 | | Structure of human HCN4 hyperpolarization-activated cyclic nucleotide-gated ion channel in complex with cAMP | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOINOSITOL, 1,2-Distearoyl-sn-glycerophosphoethanolamine, ... | | Authors: | Shintre, C.A, Pike, A.C.W, Tessitore, A, Young, M, Bushell, S.R, Strain-Damerell, C, Mukhopadhyay, S, Burgess-Brown, N.A, Huiskonen, J.T, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-06-30 | | Release date: | 2019-05-01 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of human HCN4 hyperpolarization-activated cyclic nucleotide-gated ion channel
To Be Published
|
|
4Z0O
 
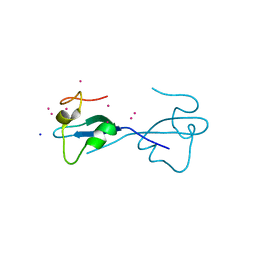 | | CW-type zinc finger of ZCWPW2 with F78D mutation | | Descriptor: | SODIUM ION, UNKNOWN ATOM OR ION, ZINC ION, ... | | Authors: | Liu, Y, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-03-26 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | CW-type zinc finger of ZCWPW2 with F78D mutation
To be Published
|
|
4ZHN
 
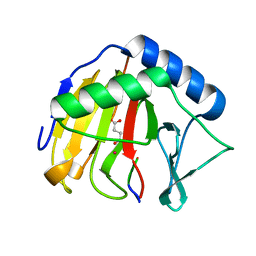 | | Crystal Structure of AlkB T208A mutant protein in complex with Co(II), 2-oxoglutarate, and methylated trinucleotide T-meA-T | | Descriptor: | 2-OXOGLUTARIC ACID, Alpha-ketoglutarate-dependent dioxygenase AlkB, COBALT (II) ION, ... | | Authors: | Ergel, B, Yu, B, Vorobiev, S.M, Forouhar, F, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-04-25 | | Release date: | 2015-12-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.333 Å) | | Cite: | DFT Studies of the Rate-Limiting Step in the Reaction Cycle of the Fe/2OG Dioxygenase AlkB and Related Experimental Studies
To be published
|
|
5BO3
 
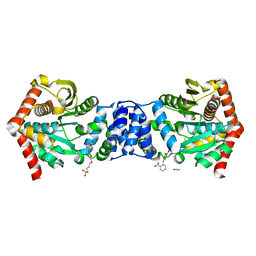 | |
4ZRA
 
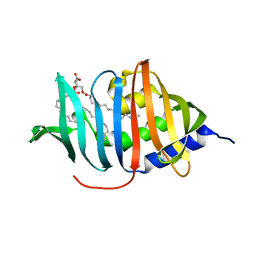 | | CRYSTAL STRUCTURE OF MYCOBACTERIUM TUBERCULOSIS LPRG BINDING TO TRIACYLGLYCERIDE | | Descriptor: | Lipoprotein LprG, Tripalmitoylglycerol | | Authors: | Martinot, A.J, Farrow, M, Bai, L, Layre, E, Cheng, T.Y, Tsai, J.H.C, Iqbal, J, Annand, J, Sullivan, Z, Hussain, M, Sacchettini, J, Moody, D.B, Seeliger, J, Rubin, E.J, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2015-05-12 | | Release date: | 2016-02-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Mycobacterial Metabolic Syndrome: LprG and Rv1410 Regulate Triacylglyceride Levels, Growth Rate and Virulence in Mycobacterium tuberculosis.
Plos Pathog., 12, 2016
|
|
1LXJ
 
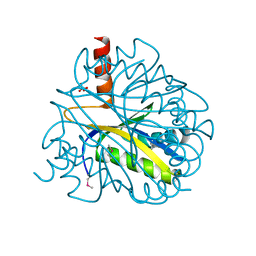 | | X-RAY STRUCTURE OF YBL001c NORTHEAST STRUCTURAL GENOMICS (NESG) CONSORTIUM TARGET YTYst72 | | Descriptor: | HYPOTHETICAL 11.5KDA PROTEIN IN HTB2-NTH2 INTERGENIC REGION, SULFATE ION | | Authors: | Tao, X, Khayat, R, Christendat, D, Savchenko, A, Xu, X, Edwards, A, Arrowsmith, C.H, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-06-05 | | Release date: | 2003-07-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | CRYSTAL STRUCTURES OF MTH1187 AND ITS YEAST ORTHOLOG YBL001C
Proteins, 52, 2003
|
|
4FPW
 
 | | Crystal Structure of CalU16 from Micromonospora echinospora. Northeast Structural Genomics Consortium Target MiR12. | | Descriptor: | CalU16 | | Authors: | Seetharaman, J, Lew, S, Janjua, H, Xiao, R, Acton, T.B, Everett, J.K, Phillips Jr, G.N, Kennedy, M.A, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-06-22 | | Release date: | 2012-12-12 | | Last modified: | 2014-11-12 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Guided Functional Characterization of Enediyne Self-Sacrifice Resistance Proteins, CalU16 and CalU19.
Acs Chem.Biol., 9, 2014
|
|
3MGJ
 
 | | Crystal structure of the Saccharop_dh_N domain of MJ1480 protein from Methanococcus jannaschii. Northeast Structural Genomics Consortium Target MjR83a. | | Descriptor: | Uncharacterized protein MJ1480 | | Authors: | Vorobiev, S, Neely, H, Seetharaman, J, Lee, D, Patel, D, Ciccosanti, C, Xiao, R, Acton, T.B, Everett, J.K, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-04-06 | | Release date: | 2010-04-21 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Crystal structure of the Saccharop_dh_N domain of MJ1480 protein from Methanococcus jannaschii.
To be Published
|
|
3NLC
 
 | | Crystal structure of the VP0956 protein from Vibrio parahaemolyticus. Northeast Structural Genomics Consortium Target VpR147 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Uncharacterized protein VP0956 | | Authors: | Vorobiev, S, Neely, H, Seetharaman, J, Wang, D, Mao, L, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-06-21 | | Release date: | 2010-08-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of the VP0956 protein from Vibrio parahaemolyticus.
To be Published
|
|
3O6U
 
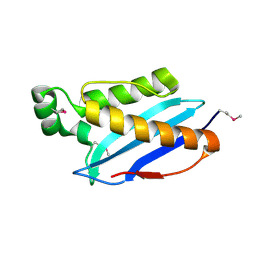 | | Crystal Structure of CPE2226 protein from Clostridium perfringens. Northeast Structural Genomics Consortium Target CpR195 | | Descriptor: | uncharacterized protein CPE2226 | | Authors: | Vorobiev, S, Su, M, Seetharaman, J, Patel, P, Xiao, R, Ciccosanti, C, Wang, H, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-07-29 | | Release date: | 2010-08-11 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of CPE2226 protein from Clostridium perfringens.
To be Published
|
|
6Y2D
 
 | | Crystal structure of the second KH domain of FUBP1 | | Descriptor: | Far upstream element-binding protein 1, GLYCEROL, SULFATE ION | | Authors: | Ni, X, Chaikuad, A, Joerger, A.C, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-02-15 | | Release date: | 2020-03-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Comparative structural analyses and nucleotide-binding characterization of the four KH domains of FUBP1.
Sci Rep, 10, 2020
|
|
6Y2C
 
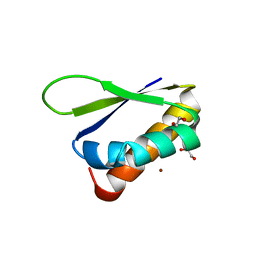 | | Crystal structure of the third KH domain of FUBP1 | | Descriptor: | 1,2-ETHANEDIOL, Far upstream element-binding protein 1, ZINC ION | | Authors: | Ni, X, Joerger, A.C, Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-02-15 | | Release date: | 2020-03-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Comparative structural analyses and nucleotide-binding characterization of the four KH domains of FUBP1.
Sci Rep, 10, 2020
|
|
3NNG
 
 | | Crystal structure of the F5/8 type C domain of Q5LFR2_BACFN protein from Bacteroides fragilis. Northeast Structural Genomics Consortium Target BfR258E | | Descriptor: | CALCIUM ION, uncharacterized protein | | Authors: | Vorobiev, S, Su, M, Dimaio, F, Baker, D, Seetharaman, J, Janjua, J, Xiao, R, Ciccosanti, C, Foote, E.L, Lee, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-06-23 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.177 Å) | | Cite: | Crystal structure of the F5/8 type C domain of Q5LFR2_BACFN protein from Bacteroides fragilis.
To be Published
|
|
3NJC
 
 | | Crystal structure of the yslB protein from Bacillus subtilis. Northeast Structural Genomics Consortium Target SR460. | | Descriptor: | YSLB protein | | Authors: | Vorobiev, S, Abashidze, M, Seetharaman, J, Wang, D, Cunningham, K, Ma, L.-C, Owens, L, Fang, Y, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-06-17 | | Release date: | 2010-09-22 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.693 Å) | | Cite: | Crystal structure of the yslB protein from Bacillus subtilis.
To be Published
|
|
3PU2
 
 | | Crystal Structure of the Q3J4M4_RHOS4 protein from Rhodobacter sphaeroides. Northeast Structural Genomics Consortium Target RhR263. | | Descriptor: | uncharacterized protein | | Authors: | Vorobiev, S, Chen, Y, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Wang, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-12-03 | | Release date: | 2010-12-15 | | Last modified: | 2012-02-22 | | Method: | X-RAY DIFFRACTION (2.606 Å) | | Cite: | Crystal Structure of the Q3J4M4_RHOS4 protein from Rhodobacter sphaeroides.
To be Published
|
|
5HTC
 
 | | Crystal structure of haspin (GSG2) in complex with bisubstrate inhibitor ARC-3372 | | Descriptor: | (2R)-2-{[6-({[(2S,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]carbonyl}amino)hexanoyl]amino}butanedioic acid (non-preferred name), (4S)-2-METHYL-2,4-PENTANEDIOL, ARC-3372 INHIBITOR, ... | | Authors: | Chaikuad, A, Heroven, C, Lavogina, D, Kestav, K, Uri, A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-01-26 | | Release date: | 2016-03-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Co-crystal structures of the protein kinase haspin with bisubstrate inhibitors.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
3CNW
 
 | | Three-dimensional structure of the protein XoxI (Q81AY6) from Bacillus cereus. Northeast Structural Genomics Consortium target BcR196. | | Descriptor: | Protein XoxI | | Authors: | Kuzin, A.P, Abashidze, M, Seetharaman, J, Wang, H, Ciccosanti, C, Mao, L, Xiao, R, Nair, R, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-03-26 | | Release date: | 2008-04-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Three-dimensional structure of the protein XoxI (Q81AY6) from Bacillus cereus. Northeast Structural Genomics Consortium target BcR196.
To Be Published
|
|
3NZP
 
 | | Crystal Structure of the Biosynthetic Arginine decarboxylase SpeA from Campylobacter jejuni, Northeast Structural Genomics Consortium Target BR53 | | Descriptor: | Arginine decarboxylase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Forouhar, F, Lew, S, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Belote, R.L, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-07-16 | | Release date: | 2010-09-01 | | Last modified: | 2012-02-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of bacterial biosynthetic arginine decarboxylases.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
3NZQ
 
 | | Crystal Structure of Biosynthetic arginine decarboxylase ADC (SpeA) from Escherichia coli, Northeast Structural Genomics Consortium Target ER600 | | Descriptor: | Biosynthetic arginine decarboxylase, SULFATE ION | | Authors: | Forouhar, F, Lew, S, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Belote, R.L, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-07-16 | | Release date: | 2010-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of bacterial biosynthetic arginine decarboxylases.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
3NNQ
 
 | | Crystal Structure of the N-terminal domain of Moloney murine leukemia virus integrase, Northeast Structural Genomics Consortium Target OR3 | | Descriptor: | ACETATE ION, N-terminal domain of Moloney murine leukemia virus integrase, ZINC ION | | Authors: | Guan, R, Xiao, R, Acton, T, Jiang, M, Roth, M, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-06-24 | | Release date: | 2010-07-14 | | Last modified: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (2.693 Å) | | Cite: | X-ray crystal structure of the N-terminal region of Moloney murine leukemia virus integrase and its implications for viral DNA recognition.
Proteins, 85, 2017
|
|
1JRM
 
 | |
5FE5
 
 | | Crystal structure of human PCAF bromodomain in complex with fragment MB093 (fragment 7) | | Descriptor: | 1,2-ETHANEDIOL, 1-[4-(1,2,3-thiadiazol-4-yl)phenyl]methanamine, DIMETHYL SULFOXIDE, ... | | Authors: | Chaikuad, A, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-12-16 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structure-Based Identification of Inhibitory Fragments Targeting the p300/CBP-Associated Factor Bromodomain.
J.Med.Chem., 59, 2016
|
|
5FE4
 
 | | Crystal structure of human PCAF bromodomain in complex with fragment MB364 (fragment 5) | | Descriptor: | 1,2-ETHANEDIOL, 2,3-dihydro-1,4-benzodioxine-5-carboxamide, Histone acetyltransferase KAT2B | | Authors: | Chaikuad, A, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-12-16 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure-Based Identification of Inhibitory Fragments Targeting the p300/CBP-Associated Factor Bromodomain.
J.Med.Chem., 59, 2016
|
|
5FE1
 
 | | Crystal structure of human PCAF bromodomain in complex with fragment BR004 (fragment 1) | | Descriptor: | 1,2-ETHANEDIOL, 1-METHYLQUINOLIN-2(1H)-ONE, Histone acetyltransferase KAT2B | | Authors: | Chaikuad, A, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-12-16 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structure-Based Identification of Inhibitory Fragments Targeting the p300/CBP-Associated Factor Bromodomain.
J.Med.Chem., 59, 2016
|
|
5FE8
 
 | | Crystal structure of human PCAF bromodomain in complex with compound SL1126 (compound 12) | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, Histone acetyltransferase KAT2B, ... | | Authors: | Chaikuad, A, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-12-16 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Identification of Inhibitory Fragments Targeting the p300/CBP-Associated Factor Bromodomain.
J.Med.Chem., 59, 2016
|
|
