1ULR
 
 | | Crystal structure of tt0497 from Thermus thermophilus HB8 | | Descriptor: | putative acylphosphatase | | Authors: | Ago, H, Hamada, K, Sugahara, M, Kuroishi, C, Kuramitsu, S, Yokoyama, S, Miyano, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-09-16 | | Release date: | 2004-11-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of tt0497 from Thermus thermophilus HB8
To be Published
|
|
1UMD
 
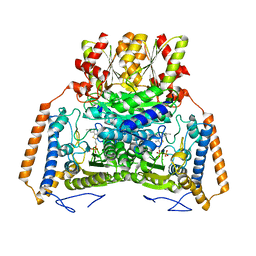 | | branched-chain 2-oxo acid dehydrogenase (E1) from Thermus thermophilus HB8 with 4-methyl-2-oxopentanoate as an intermediate | | Descriptor: | 2-OXO-4-METHYLPENTANOIC ACID, 2-oxo acid dehydrogenase alpha subunit, 2-oxo acid dehydrogenase beta subunit, ... | | Authors: | Nakai, T, Nakagawa, N, Maoka, N, Masui, R, Kuramitsu, S, Kamiya, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-09-25 | | Release date: | 2004-03-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ligand-induced Conformational Changes and a Reaction Intermediate in Branched-chain 2-Oxo Acid Dehydrogenase (E1) from Thermus thermophilus HB8, as Revealed by X-ray Crystallography
J.Mol.Biol., 337, 2004
|
|
1V27
 
 | |
2PA0
 
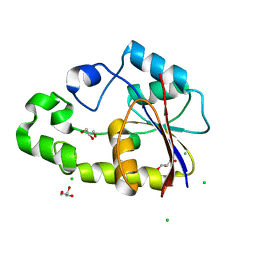 | | Crystal structure of TTHB049 from Thermus thermophilus HB8 | | Descriptor: | Alpha-ribazole-5'-phosphate phosphatase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Sugahara, M, Tanaka, Y, Matsuura, Y, Nakamoto, T, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-27 | | Release date: | 2007-10-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of TTHB049 from Thermus thermophilus HB8
To be Published
|
|
2PBR
 
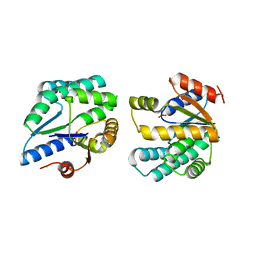 | | Crystal structure of thymidylate kinase (aq_969) from Aquifex Aeolicus VF5 | | Descriptor: | SULFATE ION, Thymidylate kinase | | Authors: | Jeyakanthan, J, Kanaujia, S.P, Vasuki Ranjani, C, Sekar, K, Nakagawa, N, Ebihara, A, Kuramitsu, S, Shinkai, A, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-29 | | Release date: | 2007-10-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal structure of thymidylate kinase (aq_969) from Aquifex Aeolicus VF5
To be Published
|
|
1V38
 
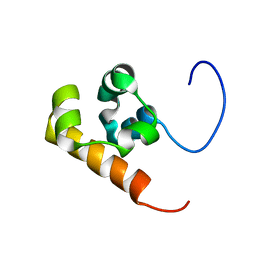 | | Solution structure of the Sterile Alpha Motif (SAM) domain of mouse SAMSN1 | | Descriptor: | SAM-domain protein SAMSN-1 | | Authors: | Goroncy, A, Kigawa, T, Koshiba, S, Kobayashi, N, Tochio, N, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-10-29 | | Release date: | 2004-04-29 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Sterile Alpha Motif (SAM) domain of mouse SAMSN1
To be Published
|
|
1V4P
 
 | |
2PCN
 
 | | Crystal structure of S-adenosylmethionine: 2-dimethylmenaquinone methyltransferase (gk_1813) from geobacillus kaustophilus HTA426 | | Descriptor: | ACETATE ION, S-adenosylmethionine:2-demethylmenaquinone methyltransferase | | Authors: | Jeyakanthan, J, Kanaujia, S.P, Rafi, Z.A, Sekar, K, Agari, Y, Ebihara, A, Kuramitsu, S, Shinkai, A, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-30 | | Release date: | 2007-10-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of S-adenosylmethionine:2-dimethylmenaquinone methyltransferase (gk_1813) from geobacillus kaustophilus HTA426
To be Published
|
|
1V7Q
 
 | |
1V93
 
 | | 5,10-Methylenetetrahydrofolate Reductase from Thermus thermophilus HB8 | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 5,10-Methylenetetrahydrofolate reductase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Nakajima, Y, Miyahara, I, Hirotsu, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-01-20 | | Release date: | 2004-02-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of 5,10-Methylenetetrahydrofolate reductase from Thermus thermophilus HB8
To be Published
|
|
1UMB
 
 | | branched-chain 2-oxo acid dehydrogenase (E1) from Thermus thermophilus HB8 in holo-form | | Descriptor: | 2-oxo acid dehydrogenase alpha subunit, 2-oxo acid dehydrogenase beta subunit, MAGNESIUM ION, ... | | Authors: | Nakai, T, Nakagawa, N, Maoka, N, Masui, R, Kuramitsu, S, Kamiya, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-09-25 | | Release date: | 2004-03-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Ligand-induced Conformational Changes and a Reaction Intermediate in Branched-chain 2-Oxo Acid Dehydrogenase (E1) from Thermus thermophilus HB8, as Revealed by X-ray Crystallography
J.Mol.Biol., 337, 2004
|
|
1V3W
 
 | | Structure of Ferripyochelin binding protein from Pyrococcus horikoshii OT3 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Jeyakanthan, J, Tahirov, T.H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-07 | | Release date: | 2003-11-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Observation of a calcium-binding site in the gamma-class carbonic anhydrase from Pyrococcus horikoshii.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
1V47
 
 | | Crystal structure of ATP sulfurylase from Thermus thermophillus HB8 in complex with APS | | Descriptor: | ADENOSINE-5'-PHOSPHOSULFATE, ATP sulfurylase, CHLORIDE ION, ... | | Authors: | Taguchi, Y, Sugishima, M, Fukuyama, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-11 | | Release date: | 2004-04-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Crystal structure of a novel zinc-binding ATP sulfurylase from Thermus thermophilus HB8
Biochemistry, 43, 2004
|
|
1V5V
 
 | |
1V8E
 
 | |
1V9S
 
 | |
1VA9
 
 | |
1VC3
 
 | |
1V6G
 
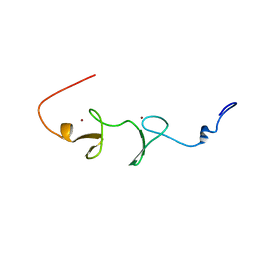 | | Solution Structure of the LIM Domain of the Human Actin Binding LIM Protein 2 | | Descriptor: | Actin Binding LIM Protein 2, ZINC ION | | Authors: | Miyamoto, K, Tomizawa, T, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-29 | | Release date: | 2004-06-01 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the LIM Domain of the Human Actin Binding LIM Protein 2
To be Published
|
|
2P79
 
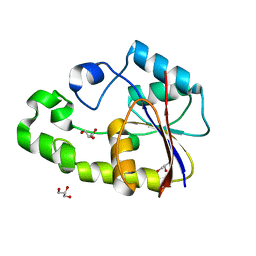 | | Crystal structure of TTHB049 from Thermus thermophilus HB8 | | Descriptor: | Alpha-ribazole-5'-phosphate phosphatase, GLYCEROL, SODIUM ION | | Authors: | Sugahara, M, Taketa, M, Kageyama, Y, Matsuura, Y, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-20 | | Release date: | 2007-09-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of TTHB049 from Thermus thermophilus HB8
To be Published
|
|
1V95
 
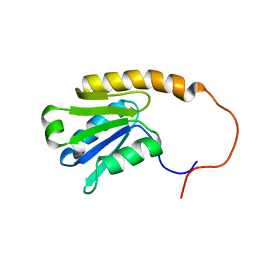 | | Solution Structure of Anticodon Binding Domain from Nuclear Receptor Coactivator 5 (Human KIAA1637 Protein) | | Descriptor: | Nuclear receptor coactivator 5 | | Authors: | Zhao, C, Kigawa, T, Tochio, N, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-01-20 | | Release date: | 2004-07-20 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Anticodon Binding Domain from Nuclear Receptor Coactivator 5 (Human KIAA1637 Protein)
To be Published
|
|
1VCD
 
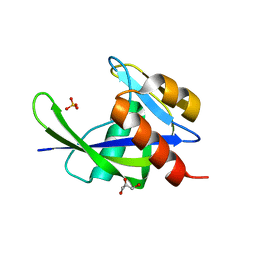 | | Crystal Structure of a T.thermophilus HB8 Ap6A hydrolase Ndx1 | | Descriptor: | GLYCEROL, Ndx1, SULFATE ION | | Authors: | Iwai, T, Nakagawa, N, Kuramitsu, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-03-05 | | Release date: | 2005-04-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Nudix Protein Ndx1 from Thermus thermophilus HB8 in binary complex with diadenosine hexaphosphate
To be Published
|
|
2P9F
 
 | | Crystal structure of TTHB049 from Thermus thermophilus HB8 | | Descriptor: | Alpha-ribazole-5'-phosphate phosphatase, GLYCEROL | | Authors: | Yamamoto, H, Taketa, M, Kageyama, Y, Matsuura, Y, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-25 | | Release date: | 2007-09-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of TTHB049 from Thermus thermophilus HB8
To be Published
|
|
1UM7
 
 | |
1V2D
 
 | |
