5F96
 
 | |
4U0I
 
 | | Crystal structure of KIT in complex with ponatinib | | Descriptor: | 3-(imidazo[1,2-b]pyridazin-3-ylethynyl)-4-methyl-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}benzam ide, Mast/stem cell growth factor receptor Kit,Mast/stem cell growth factor receptor Kit, PHOSPHATE ION | | Authors: | Zhou, T, Zhu, X, Dalgarno, D.C. | | Deposit date: | 2014-07-11 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ponatinib Inhibits Polyclonal Drug-Resistant KIT Oncoproteins and Shows Therapeutic Potential in Heavily Pretreated Gastrointestinal Stromal Tumor (GIST) Patients.
Clin.Cancer Res., 20, 2014
|
|
7TCQ
 
 | |
7MXD
 
 | |
7N28
 
 | |
5V7J
 
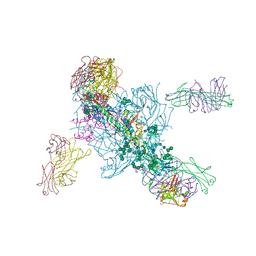 | | Crystal Structure at 3.7 A Resolution of Glycosylated HIV-1 Clade A BG505 SOSIP.664 Prefusion Env Trimer with Four Glycans (N197, N276, N362, and N462) removed in Complex with Neutralizing Antibodies 3H+109L and 35O22. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody 35O22 Fab heavy chain, ... | | Authors: | Stewart-Jones, G.B.E, Zhou, T, Kwong, P.D. | | Deposit date: | 2017-03-20 | | Release date: | 2017-06-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.907 Å) | | Cite: | Quantification of the Impact of the HIV-1-Glycan Shield on Antibody Elicitation.
Cell Rep, 19, 2017
|
|
5EQE
 
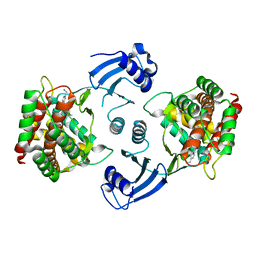 | | Crystal structure of choline kinase alpha-1 bound by [4-[(4-methyl-1,4-diazepan-1-yl)methyl]phenyl]methanamine (compound 11) | | Descriptor: | Choline kinase alpha, [4-[(4-methyl-1,4-diazepan-1-yl)methyl]phenyl]methanamine | | Authors: | Zhou, T, Zhu, X, Dalgarno, D.C. | | Deposit date: | 2015-11-12 | | Release date: | 2016-01-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Novel Small Molecule Inhibitors of Choline Kinase Identified by Fragment-Based Drug Discovery.
J.Med.Chem., 59, 2016
|
|
5EQP
 
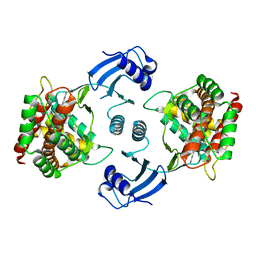 | | Crystal structure of choline kinase alpha-1 bound by 6-[(4-methyl-1,4-diazepan-1-yl)methyl]quinoline (compound 37) | | Descriptor: | 6-[(4-methyl-1,4-diazepan-1-yl)methyl]quinoline, Choline kinase alpha | | Authors: | Zhou, T, Zhu, X, Dalgarno, D.C. | | Deposit date: | 2015-11-13 | | Release date: | 2016-01-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Novel Small Molecule Inhibitors of Choline Kinase Identified by Fragment-Based Drug Discovery.
J.Med.Chem., 59, 2016
|
|
5EQY
 
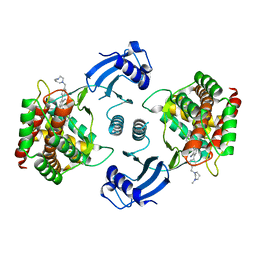 | | Crystal structure of choline kinase alpha-1 bound by 5-[(4-methyl-1,4-diazepan-1-yl)methyl]-2-[4-[(4-methyl-1,4-diazepan-1-yl)methyl]phenyl]benzenecarbonitrile (compound 65) | | Descriptor: | 5-[(4-methyl-1,4-diazepan-1-yl)methyl]-2-[4-[(4-methyl-1,4-diazepan-1-yl)methyl]phenyl]benzenecarbonitrile, Choline kinase alpha | | Authors: | Zhou, T, Zhu, X, Dalgarno, D.C. | | Deposit date: | 2015-11-13 | | Release date: | 2016-01-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Novel Small Molecule Inhibitors of Choline Kinase Identified by Fragment-Based Drug Discovery.
J.Med.Chem., 59, 2016
|
|
5JOE
 
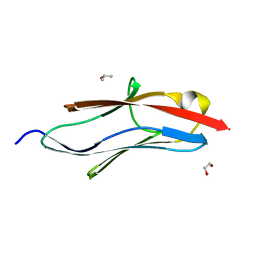 | | Crystal structure of I81 from titin | | Descriptor: | 1,2-ETHANEDIOL, ISOPROPYL ALCOHOL, Titin | | Authors: | Fleming, J, Zhou, T, Bogomolovas, J, Labeit, S, Mayans, O. | | Deposit date: | 2016-05-02 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CARP interacts with titin at a unique helical N2A sequence and at the domain Ig81 to form a structured complex.
Febs Lett., 590, 2016
|
|
8TOP
 
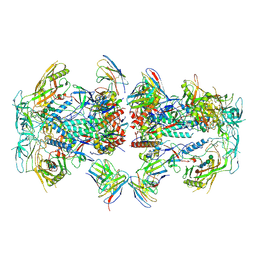 | | Cryo-EM structure of HIV-1 Env BG505 DS-SOSIP in complex with antibody GPZ6-b.01 targeting the fusion peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HIV-1 BG505 DS-SOSIP glycoprotein gp41, ... | | Authors: | Zhou, T, Morano, N.C, Roark, R.S, Kwong, P.D. | | Deposit date: | 2023-08-03 | | Release date: | 2024-08-07 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Potent and broad HIV-1 neutralization in fusion peptide-primed SHIV-infected macaques.
Cell, 187, 2024
|
|
8TNI
 
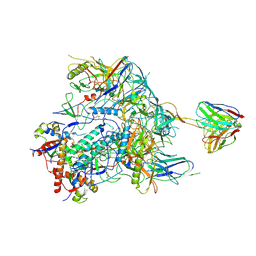 | | Cryo-EM structure of HIV-1 Env BG505 DS-SOSIP in complex with broadly neutralizing bi-specific antibody CAP256L-R27 targeting the CD4-binding site and the V2-apex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HIV-1 BG505 DS-SOSIP gp120, ... | | Authors: | Zhou, T, Morano, N.C, Roark, R.S, Kwong, P.D, Xu, J. | | Deposit date: | 2023-08-01 | | Release date: | 2024-05-08 | | Last modified: | 2024-12-18 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | Ultrapotent Broadly Neutralizing Human-llama Bispecific Antibodies against HIV-1.
Adv Sci, 11, 2024
|
|
8TNH
 
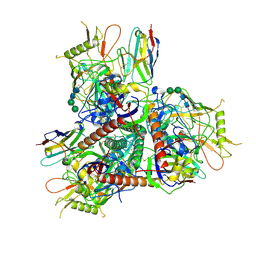 | | Cryo-EM structure of HIV-1 Env BG505 DS-SOSIP in complex with broadly neutralizing llama nanobody G36 targeting the CD4-binding site | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CD4-binding site nanobody G36, ... | | Authors: | Zhou, T, Kwong, P.D, Xu, J. | | Deposit date: | 2023-08-01 | | Release date: | 2024-05-08 | | Last modified: | 2024-12-18 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Ultrapotent Broadly Neutralizing Human-llama Bispecific Antibodies against HIV-1.
Adv Sci, 11, 2024
|
|
8TNG
 
 | | Cryo-EM structure of HIV-1 Env BG505 DS-SOSIP in complex with broadly neutralizing llama nanobody R27 targeting the CD4-binding site | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CD4-binding site targeting nanobody R27, ... | | Authors: | Zhou, T, Kwong, P.D, Xu, J. | | Deposit date: | 2023-08-01 | | Release date: | 2024-05-08 | | Last modified: | 2024-12-18 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Ultrapotent Broadly Neutralizing Human-llama Bispecific Antibodies against HIV-1.
Adv Sci, 11, 2024
|
|
8T1G
 
 | |
3SE9
 
 | | Crystal structure of broadly and potently neutralizing antibody VRC-PG04 in complex with HIV-1 gp120 | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kwong, P.D, Zhou, T. | | Deposit date: | 2011-06-10 | | Release date: | 2011-08-10 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing.
Science, 333, 2011
|
|
3SE8
 
 | | Crystal structure of broadly and potently neutralizing antibody VRC03 in complex with HIV-1 gp120 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kwong, P.D, Zhou, T. | | Deposit date: | 2011-06-10 | | Release date: | 2011-08-10 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.895 Å) | | Cite: | Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing.
Science, 333, 2011
|
|
4D49
 
 | | Crystal structure of computationally designed armadillo repeat proteins for modular peptide recognition. | | Descriptor: | ARGININE, ARMADILLO REPEAT PROTEIN ARM00027, POLY ARG DECAPEPTIDE | | Authors: | Reichen, C, Forzani, C, Zhou, T, Parmeggiani, F, Fleishman, S.J, Mittl, P.R.E, Madhurantakam, C, Honegger, A, Ewald, C, Zerbe, O, Baker, D, Caflisch, A, Pluckthun, A. | | Deposit date: | 2014-10-27 | | Release date: | 2016-01-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Computationally Designed Armadillo Repeat Proteins for Modular Peptide Recognition.
J.Mol.Biol., 428, 2016
|
|
4D4E
 
 | | Crystal structure of computationally designed armadillo repeat proteins for modular peptide recognition. | | Descriptor: | ARMADILLO REPEAT PROTEIN ARM00016, GLYCEROL | | Authors: | Reichen, C, Forzani, C, Zhou, T, Parmeggiani, F, Fleishman, S.J, Mittl, P.R.E, Madhurantakam, C, Honegger, A, Ewald, C, Zerbe, O, Baker, D, Caflisch, A, Pluckthun, A. | | Deposit date: | 2014-10-28 | | Release date: | 2016-01-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Computationally Designed Armadillo Repeat Proteins for Modular Peptide Recognition.
J.Mol.Biol., 428, 2016
|
|
3JWD
 
 | | Structure of HIV-1 gp120 with gp41-Interactive Region: Layered Architecture and Basis of Conformational Mobility | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FAB 48D HEAVY CHAIN, FAB 48D LIGHT CHAIN, ... | | Authors: | Pancera, M, Majeed, S, Ban, Y.A, Chen, L, Huang, C.C, Kong, L, Kwon, Y.D, Stuckey, J, Zhou, T, Robinson, J.E, Schief, W.R, Sodroski, J, Wyatt, R, Kwong, P.D. | | Deposit date: | 2009-09-18 | | Release date: | 2009-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structure of HIV-1 gp120 with gp41-interactive region reveals layered envelope architecture and basis of conformational mobility.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
1H74
 
 | | CRYSTAL STRUCTURE OF HOMOSERINE KINASE COMPLEXED WITH ILE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, HOMOSERINE KINASE, ISOLEUCINE, ... | | Authors: | Krishna, S.S, Zhou, T, Daugherty, M, Osterman, A.L, Zhang, H. | | Deposit date: | 2001-07-02 | | Release date: | 2001-11-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for the Catalysis and Substrate Specificity of Homoserine Kinase
Biochemistry, 40, 2001
|
|
1H72
 
 | | CRYSTAL STRUCTURE OF HOMOSERINE KINASE COMPLEXED WITH HSE | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, HOMOSERINE KINASE, L-HOMOSERINE, ... | | Authors: | Krishna, S.S, Zhou, T, Daugherty, M, Osterman, A.L, Zhang, H. | | Deposit date: | 2001-07-02 | | Release date: | 2001-09-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for the Catalysis and Substrate Specificity of Homoserine Kinase
Biochemistry, 40, 2001
|
|
1H73
 
 | | CRYSTAL STRUCTURE OF HOMOSERINE KINASE COMPLEXED WITH THREONINE | | Descriptor: | HOMOSERINE KINASE, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, THREONINE | | Authors: | Krishna, S.S, Zhou, T, Daugherty, M, Osterman, A.L, Zhang, H. | | Deposit date: | 2001-07-02 | | Release date: | 2001-09-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for the Catalysis and Substrate Specificity of Homoserine Kinase
Biochemistry, 40, 2001
|
|
4JPW
 
 | | Crystal structure of broadly and potently neutralizing antibody 12a21 in complex with hiv-1 strain 93th057 gp120 mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HEAVY CHAIN OF ANTIBODY 12A21, ... | | Authors: | Acharya, P, Luongo, T, Zhou, T, Kwong, P.D. | | Deposit date: | 2013-03-19 | | Release date: | 2013-04-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.904 Å) | | Cite: | Somatic mutations of the immunoglobulin framework are generally required for broad and potent HIV-1 neutralization.
Cell(Cambridge,Mass.), 153, 2013
|
|
5M29
 
 | | Structure of cobinamide-bound BtuF, the periplasmic vitamin B12 binding protein in E.coli | | Descriptor: | COB(II)INAMIDE, CYANIDE ION, GLYCEROL, ... | | Authors: | Mireku, S.A, Ruetz, M, Zhou, T, Korkhov, V.M, Kraeutler, B, Locher, K.P. | | Deposit date: | 2016-10-12 | | Release date: | 2017-02-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Conformational Change of a Tryptophan Residue in BtuF Facilitates Binding and Transport of Cobinamide by the Vitamin B12 Transporter BtuCD-F.
Sci Rep, 7, 2017
|
|
