7LA3
 
 | | Pseudomonas fluorescens G150A isocyanide hydratase (G150A-3) at 274K, Refmac5-refined | | Descriptor: | Isonitrile hydratase InhA | | Authors: | Su, Z, Dasgupta, M, Poitevin, F, Mathews, I.I, van den Bedem, H, Wall, M.E, Yoon, C.H, Wilson, M.A. | | Deposit date: | 2021-01-05 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.349 Å) | | Cite: | Reproducibility of protein x-ray diffuse scattering and potential utility for modeling atomic displacement parameters.
Struct Dyn., 8, 2021
|
|
7LAV
 
 | | Pseudomonas fluorescens G150T isocyanide hydratase (G150T-1) at 274K, Refmac5-refined | | Descriptor: | Isonitrile hydratase InhA | | Authors: | Su, Z, Dasgupta, M, Poitevin, F, Mathews, I.I, van den Bedem, H, Wall, M.E, Yoon, C.H, Wilson, M.A. | | Deposit date: | 2021-01-06 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.149 Å) | | Cite: | Reproducibility of protein x-ray diffuse scattering and potential utility for modeling atomic displacement parameters.
Struct Dyn., 8, 2021
|
|
7LAX
 
 | | Pseudomonas fluorescens G150T isocyanide hydratase (G150T-2) at 274K, Refmac5-refined | | Descriptor: | Isonitrile hydratase InhA | | Authors: | Su, Z, Dasgupta, M, Poitevin, F, Mathews, I.I, van den Bedem, H, Wall, M.E, Yoon, C.H, Wilson, M.A. | | Deposit date: | 2021-01-07 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.198 Å) | | Cite: | Reproducibility of protein x-ray diffuse scattering and potential utility for modeling atomic displacement parameters.
Struct Dyn., 8, 2021
|
|
7LB9
 
 | | Pseudomonas fluorescens G150T isocyanide hydratase (G150T-3) at 274K, Refmac5-refined | | Descriptor: | Isonitrile hydratase InhA | | Authors: | Su, Z, Dasgupta, M, Poitevin, F, Mathews, I.I, van den Bedem, H, Wall, M.E, Yoon, C.H, Wilson, M.A. | | Deposit date: | 2021-01-07 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.101 Å) | | Cite: | Reproducibility of protein x-ray diffuse scattering and potential utility for modeling atomic displacement parameters.
Struct Dyn., 8, 2021
|
|
7LBI
 
 | | Wild-type Pseudomonas fluorescens isocyanide hydratase (WT-2) at 274K, PHENIX-refined | | Descriptor: | Isonitrile hydratase InhA | | Authors: | Su, Z, Dasgupta, M, Poitevin, F, Mathews, I.I, van den Bedem, H, Wall, M.E, Yoon, C.H, Wilson, M.A. | | Deposit date: | 2021-01-08 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Reproducibility of protein x-ray diffuse scattering and potential utility for modeling atomic displacement parameters.
Struct Dyn., 8, 2021
|
|
7LD7
 
 | | G150A Pseudomonas fluorescens isocyanide hydratase (G150A-2) at 274K, Phenix-refined | | Descriptor: | Isonitrile hydratase InhA | | Authors: | Su, Z, Dasgupta, M, Yoon, C.H, Wilson, M.A. | | Deposit date: | 2021-01-12 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Reproducibility of protein x-ray diffuse scattering and potential utility for modeling atomic displacement parameters.
Struct Dyn., 8, 2021
|
|
7LDI
 
 | | G150T Pseudomonas fluorescens isocyanide hydratase (G150T-2) at 274K, Phenix-refined | | Descriptor: | Isonitrile hydratase InhA | | Authors: | Su, Z, Dasgupta, M, Yoon, C.H, Wilson, M.A. | | Deposit date: | 2021-01-13 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Reproducibility of protein x-ray diffuse scattering and potential utility for modeling atomic displacement parameters.
Struct Dyn., 8, 2021
|
|
7LBH
 
 | | Wild-type Pseudomonas fluorescens isocyanide hydratase (WT-1) at 274K, PHENIX-refined | | Descriptor: | Isonitrile hydratase InhA | | Authors: | Su, Z, Dasgupta, M, Poitevin, F, Mathews, I.I, van den Bedem, H, Wall, M.E, Yoon, C.H, Wilson, M.A. | | Deposit date: | 2021-01-08 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Reproducibility of protein x-ray diffuse scattering and potential utility for modeling atomic displacement parameters.
Struct Dyn., 8, 2021
|
|
7LDM
 
 | | G150T Pseudomonas fluorescens isocyanide hydratase (G150T-1) at 274K, Phenix-refined | | Descriptor: | Isonitrile hydratase InhA | | Authors: | Su, Z, Dasgupta, M, Yoon, C.H, Wilson, M.A. | | Deposit date: | 2021-01-13 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Reproducibility of protein x-ray diffuse scattering and potential utility for modeling atomic displacement parameters.
Struct Dyn., 8, 2021
|
|
7LCX
 
 | | Wild-type Pseudomonas fluorescens isocyanide hydratase (WT-3) at 274K, Phenix-refined | | Descriptor: | Isonitrile hydratase InhA | | Authors: | Su, Z, Dasgupta, M, Yoon, C.H, Wilson, M.A. | | Deposit date: | 2021-01-12 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Reproducibility of protein x-ray diffuse scattering and potential utility for modeling atomic displacement parameters.
Struct Dyn., 8, 2021
|
|
7LDB
 
 | | G150A Pseudomonas fluorescens isocyanide hydratase (G150A-3) at 274K, Phenix-refined | | Descriptor: | Isonitrile hydratase InhA | | Authors: | Su, Z, Dasgupta, M, Yoon, C.H, Wilson, M.A. | | Deposit date: | 2021-01-13 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Reproducibility of protein x-ray diffuse scattering and potential utility for modeling atomic displacement parameters.
Struct Dyn., 8, 2021
|
|
1UP5
 
 | | Chicken Calmodulin | | Descriptor: | CALCIUM ION, Calmodulin | | Authors: | Wilson, M.A, Rupp, B. | | Deposit date: | 2003-09-27 | | Release date: | 2005-03-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallization and Preliminary X-Ray Analysis of Two New Crystal Forms of Calmodulin
Acta Crystallogr.,Sect.D, 52, 1996
|
|
7D49
 
 | |
7D3Z
 
 | |
7D4L
 
 | |
7D4X
 
 | |
7D6G
 
 | |
2RH6
 
 | |
8OKH
 
 | |
7YWY
 
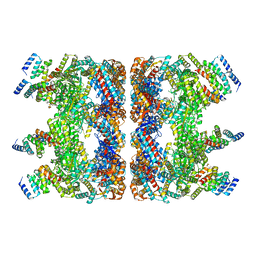 | | Structure of the GroEL chaperonin in complex with the CnoX chaperedoxin | | Descriptor: | Chaperedoxin, Chaperonin GroEL | | Authors: | Van der Verren, S.E, Remaut, H, Collet, J.F, Dupuy, E. | | Deposit date: | 2022-02-15 | | Release date: | 2023-02-22 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | A molecular device for the redox quality control of GroEL/ES substrates.
Cell, 186, 2023
|
|
6WTG
 
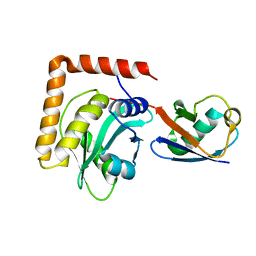 | | SdeA DUB Domain in complex with Ubiquitin | | Descriptor: | Ubiquitin, Ubiquitinating/deubiquitinating enzyme SdeA | | Authors: | Kenny, S, Sheedlo, M, Das, C. | | Deposit date: | 2020-05-02 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Insights into Ubiquitin Product Release in Hydrolysis Catalyzed by the Bacterial Deubiquitinase SdeA.
Biochemistry, 60, 2021
|
|
7T63
 
 | |
4PDJ
 
 | | Neutron crystal Structure of E.coli Dihydrofolate Reductase complexed with folate and NADP+ | | Descriptor: | DIHYDROFOLIC ACID, Dihydrofolate reductase, MANGANESE (II) ION, ... | | Authors: | Wan, Q, Kovalevsky, A.Y, Wilson, M, Langan, P, Dealwis, C, Bennett, B. | | Deposit date: | 2014-04-18 | | Release date: | 2015-04-15 | | Last modified: | 2024-04-03 | | Method: | NEUTRON DIFFRACTION (1.599 Å), X-RAY DIFFRACTION | | Cite: | Toward resolving the catalytic mechanism of dihydrofolate reductase using neutron and ultrahigh-resolution X-ray crystallography.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
8SMQ
 
 | | Crystal Structure of the N-terminal Domain of the Cryptic Surface Protein (CD630_25440) from Clostridium difficile. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Kiryukhina, O, Wawrzak, Z, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2023-04-26 | | Release date: | 2023-05-10 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Protein target highlights in CASP15: Analysis of models by structure providers.
Proteins, 91, 2023
|
|
4P3Q
 
 | | Room-temperature WT DHFR, time-averaged ensemble | | Descriptor: | CALCIUM ION, Dihydrofolate reductase, FOLIC ACID, ... | | Authors: | Keedy, D.A, van den Bedem, H, Fraser, J.S. | | Deposit date: | 2014-03-10 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.351 Å) | | Cite: | Crystal Cryocooling Distorts Conformational Heterogeneity in a Model Michaelis Complex of DHFR.
Structure, 22, 2014
|
|
