1RHX
 
 | |
1SQ8
 
 | |
1SBO
 
 | | Solution Structure of putative anti sigma factor antagonist from Thermotoga maritima (TM1442) | | Descriptor: | Putative anti-sigma factor antagonist TM1442 | | Authors: | Etezady-Esfarjaini, T, Placzek, W.J, Herrmann, T, Lesley, S.A, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2004-02-10 | | Release date: | 2004-12-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the putative anti-sigma-factor antagonist TM1442 from Thermotoga maritima in the free and phosphorylated states.
Magn.Reson.Chem., 44 Spec No, 2006
|
|
2CCX
 
 | |
1SPF
 
 | |
1QM2
 
 | |
1Z99
 
 | | Solution structure of Crotamine, a myotoxin from Crotalus durissus terrificus | | Descriptor: | Crotamine | | Authors: | Fadel, V, Bettendorff, P, Herrmann, T, de Azevedo, W.F, Oliveira, E.B, Yamane, T, Wuthrich, K. | | Deposit date: | 2005-04-01 | | Release date: | 2006-03-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Automated NMR structure determination and disulfide bond identification of the myotoxin crotamine from Crotalus durissus terrificus.
Toxicon, 46, 2005
|
|
1T6R
 
 | | Solution structure of TM1442, a putative anti sigma factor antagonist in phosphorylated state | | Descriptor: | Putative anti-sigma factor antagonist TM1442 | | Authors: | Etezady-Esfarjani, T, Placzek, W, Herrmann, T, Lesley, S.A, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2004-05-07 | | Release date: | 2005-05-24 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the putative anti-sigma-factor antagonist TM1442 from Thermotoga maritima in the free and phosphorylated states.
Magn.Reson.Chem., 44 Spec No, 2006
|
|
1GM0
 
 | | A Form of the Pheromone-Binding Protein from Bombyx mori | | Descriptor: | PHEROMONE-BINDING PROTEIN | | Authors: | Horst, R, Damberger, F, Guntert, P, Luginbuhl, P, Nikonova, L, Peng, G, Leal, W.S, Wuthrich, K. | | Deposit date: | 2001-09-05 | | Release date: | 2001-11-30 | | Last modified: | 2018-01-17 | | Method: | SOLUTION NMR | | Cite: | NMR Structure Reveals Novel Intramolecular Regulation Mechanism for Pheromone-Binding and Release
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1U3M
 
 | | NMR structure of the chicken prion protein fragment 128-242 | | Descriptor: | prion-like protein | | Authors: | Lysek, D.A, Calzolai, L, Guntert, P, Wuthrich, K. | | Deposit date: | 2004-07-22 | | Release date: | 2005-01-04 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Prion protein NMR structures of chickens, turtles, and frogs
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1H0L
 
 | |
1HA8
 
 | |
1WKT
 
 | |
1XFR
 
 | |
1XYU
 
 | | Solution structure of the sheep prion protein with polymorphism H168 | | Descriptor: | Major prion protein | | Authors: | Calzolai, L, Lysek, D.A, Guntert, P, Wuthrich, K. | | Deposit date: | 2004-11-11 | | Release date: | 2005-01-04 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Prion protein NMR structures of cats, dogs, pigs, and sheep
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
1XYK
 
 | | NMR Structure of the canine prion protein | | Descriptor: | prion protein | | Authors: | Lysek, D.A, Schorn, C, Esteve-Moya, V, Herrmann, T, Wuthrich, K. | | Deposit date: | 2004-11-10 | | Release date: | 2005-01-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Prion protein NMR structures of cats, dogs, pigs, and sheep
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
1XYQ
 
 | | NMR structure of the pig prion protein | | Descriptor: | Major prion protein | | Authors: | Lysek, D.A, Schorn, C, Herrmann, T, Wuthrich, K. | | Deposit date: | 2004-11-10 | | Release date: | 2005-01-04 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Prion protein NMR structures of cats, dogs, pigs, and sheep
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1XYW
 
 | | elk prion protein | | Descriptor: | Major prion protein | | Authors: | Gossert, A.D, Bonjour, S, Lysek, D.A, Fiorito, F, Wuthrich, K. | | Deposit date: | 2004-11-11 | | Release date: | 2004-12-28 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Prion protein NMR structures of elk and of mouse/elk hybrids
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1Y15
 
 | | Mouse Prion Protein with mutation N174T | | Descriptor: | Major prion protein | | Authors: | Gossert, A.D, Bonjour, S, Lysek, D.A, Fiorito, F, Wuthrich, K. | | Deposit date: | 2004-11-17 | | Release date: | 2004-12-28 | | Last modified: | 2021-11-10 | | Method: | SOLUTION NMR | | Cite: | Prion protein NMR structures of elk and of mouse/elk hybrids
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2FNB
 
 | | NMR STRUCTURE OF THE FIBRONECTIN ED-B DOMAIN, NMR, 20 STRUCTURES | | Descriptor: | PROTEIN (FIBRONECTIN) | | Authors: | Fattorusso, R, Pellecchia, M, Viti, F, Neri, P, Neri, D, Wuthrich, K. | | Deposit date: | 1998-12-16 | | Release date: | 1998-12-23 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the human oncofoetal fibronectin ED-B domain, a specific marker for angiogenesis.
Structure Fold.Des., 7, 1999
|
|
2GA7
 
 | | Solution structure of the copper(I) form of the third metal-binding domain of ATP7A protein (menkes disease protein) | | Descriptor: | COPPER (I) ION, Copper-transporting ATPase 1 | | Authors: | Banci, L, Bertini, I, Cantini, F, DellaMalva, N, Rosato, A, Herrmann, T, Wuthrich, K, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2006-03-08 | | Release date: | 2006-08-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure and intermolecular interactions of the third metal-binding domain of ATP7A, the Menkes disease protein.
J.Biol.Chem., 281, 2006
|
|
2G9O
 
 | | Solution structure of the apo form of the third metal-binding domain of ATP7A protein (Menkes Disease protein) | | Descriptor: | Copper-transporting ATPase 1 | | Authors: | Banci, L, Bertini, I, Cantini, F, DellaMalva, N, Rosato, A, Herrmann, T, Wuthrich, K, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2006-03-07 | | Release date: | 2006-08-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure and intermolecular interactions of the third metal-binding domain of ATP7A, the Menkes disease protein.
J.Biol.Chem., 281, 2006
|
|
2GRI
 
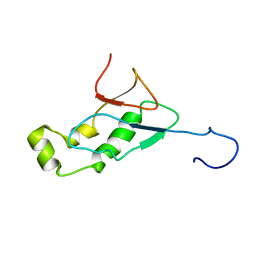 | | NMR Structure of the SARS-CoV non-structural protein nsp3a | | Descriptor: | NSP3 | | Authors: | Serrano, P, Almeida, M.S, Johnson, M.A, Herrmann, T, Saikatendu, K.S, Joseph, J, Subramanian, V, Neuman, B.W, Buchmeier, M.J, Stevens, R.C, Kuhn, P, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2006-04-24 | | Release date: | 2006-12-19 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structure of the N-terminal domain of nonstructural protein 3 from the severe acute respiratory syndrome coronavirus.
J.Virol., 81, 2007
|
|
2I3B
 
 | |
2PWT
 
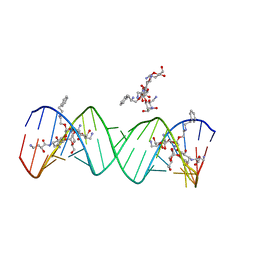 | | Crystal structure of the bacterial ribosomal decoding site complexed with aminoglycoside containing the L-HABA group | | Descriptor: | 22-mer of the ribosomal decoding site, DOUBLY FUNCTIONALIZED PAROMOMYCIN PM-II-162 | | Authors: | Kondo, J, Pachamuthu, K, Francois, B, Szychowski, J, Hanessian, S, Westhof, E. | | Deposit date: | 2007-05-13 | | Release date: | 2007-09-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the Bacterial Ribosomal Decoding Site Complexed with a Synthetic Doubly Functionalized Paromomycin Derivative: a New Specific Binding Mode to an A-Minor Motif Enhances in vitro Antibacterial Activity
Chemmedchem, 2, 2007
|
|
