7YMD
 
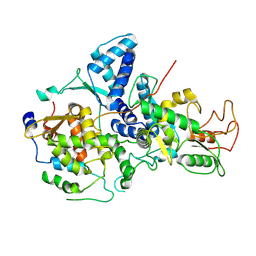 | | Cryo-EM structure of Nse1/3/4 | | Descriptor: | Non-structural maintenance of chromosome element 3, Non-structural maintenance of chromosome element 4, Non-structural maintenance of chromosomes element 1 | | Authors: | Qian, L, Jun, Z, Zhenguo, C, Wang, L. | | Deposit date: | 2022-07-28 | | Release date: | 2024-01-31 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.176 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 2024
|
|
2XAT
 
 | |
2LNL
 
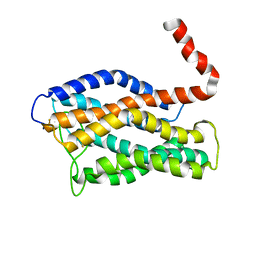 | | Structure of human CXCR1 in phospholipid bilayers | | Descriptor: | C-X-C chemokine receptor type 1 | | Authors: | Park, S, Das, B.B, Casagrande, F, Nothnagel, H, Chu, M, Kiefer, H, Maier, K, De Angelis, A, Marassi, F.M, Opella, S.J. | | Deposit date: | 2011-12-31 | | Release date: | 2012-10-17 | | Last modified: | 2016-04-27 | | Method: | SOLID-STATE NMR | | Cite: | Structure of the chemokine receptor CXCR1 in phospholipid bilayers.
Nature, 491, 2012
|
|
6IY3
 
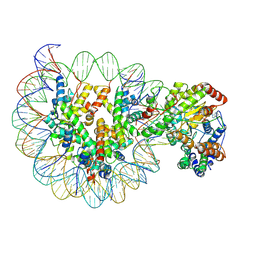 | | Structure of Snf2-MMTV-A nucleosome complex at shl-2 in ADP state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (147-MER), Histone H2A, ... | | Authors: | Li, M, Xia, X, Liu, X, Li, X, Chen, Z. | | Deposit date: | 2018-12-12 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Mechanism of DNA translocation underlying chromatin remodelling by Snf2.
Nature, 567, 2019
|
|
6IY2
 
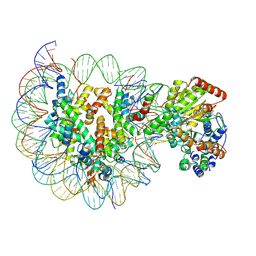 | | Structure of Snf2-MMTV-A nucleosome complex at shl2 in ADP state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (147-MER), DNA (167-MER), ... | | Authors: | Li, M, Xia, X, Liu, X, Li, X, Chen, Z. | | Deposit date: | 2018-12-12 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Mechanism of DNA translocation underlying chromatin remodelling by Snf2.
Nature, 567, 2019
|
|
1XAT
 
 | |
8WJL
 
 | | Cryo-EM structure of 6-subunit Smc5/6 hinge region | | Descriptor: | E3 SUMO-protein ligase MMS21, Structural maintenance of chromosomes protein 5, Structural maintenance of chromosomes protein 6 | | Authors: | Li, Q, Zhang, J, Zhang, X, Cheng, T, Wang, Z, Jin, D, Chen, Z, Wang, L. | | Deposit date: | 2023-09-26 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (6.15 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8WJN
 
 | | Cryo-EM structure of 6-subunit Smc5/6 head region | | Descriptor: | Non-structural maintenance of chromosome element 3, Non-structural maintenance of chromosomes element 1, Non-structural maintenance of chromosomes element 4, ... | | Authors: | Li, Q, Zhang, J, Zhang, X, Cheng, T, Wang, Z, Jin, D, Chen, Z, Wang, L. | | Deposit date: | 2023-09-26 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (5.58 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8WJO
 
 | | Cryo-EM structure of 8-subunit Smc5/6 arm region | | Descriptor: | DNA repair protein KRE29, E3 SUMO-protein ligase MMS21, Structural maintenance of chromosomes protein 5, ... | | Authors: | Li, Q, Zhang, J, Zhang, X, Cheng, T, Wang, Z, Jin, D, Chen, Z, Wang, L. | | Deposit date: | 2023-09-26 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (6.04 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 31, 2024
|
|
7X7W
 
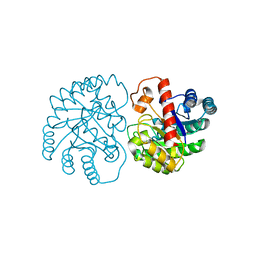 | | The X-ray Crystallographic Structure of D-Psicose 3-epimerase from Clostridia bacterium | | Descriptor: | D-PSICOSE 3-EPIMERASE, MANGANESE (II) ION | | Authors: | Li, Z.F, Ban, X.F, Xie, X.F, Tian, Y.X, Li, C.M, Gu, Z.B. | | Deposit date: | 2022-03-10 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | Crystal structure of a novel homodimeric D-allulose 3-epimerase from a Clostridia bacterium
Acta Crystallogr.,Sect.D, 78, 2022
|
|
6JXA
 
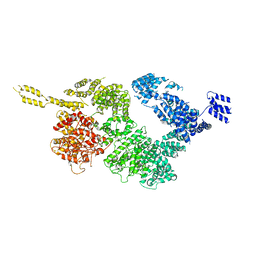 | | Tel1 kinase compact monomer | | Descriptor: | Serine/threonine-protein kinase TEL1 | | Authors: | Xin, J. | | Deposit date: | 2019-04-23 | | Release date: | 2019-07-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural basis of allosteric regulation of Tel1/ATM kinase.
Cell Res., 29, 2019
|
|
6JXC
 
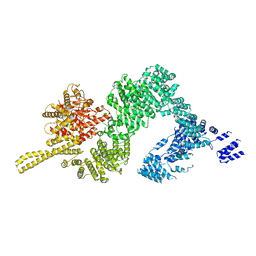 | | Tel1 kinase butterfly symmetric dimer | | Descriptor: | Serine/threonine-protein kinase TEL1 | | Authors: | Xin, J. | | Deposit date: | 2019-04-23 | | Release date: | 2019-07-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis of allosteric regulation of Tel1/ATM kinase.
Cell Res., 29, 2019
|
|
6KY5
 
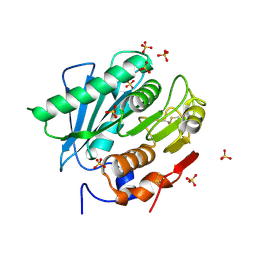 | | Crystal structure of a hydrolase mutant | | Descriptor: | PET hydrolase, SULFATE ION | | Authors: | Cui, Y.L, Chen, Y.C, Liu, X.Y, Dong, S.J, Han, J, Xiang, H, Chen, Q, Liu, H.Y, Han, X, Liu, W.D, Tang, S.Y, Wu, B. | | Deposit date: | 2019-09-16 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.631 Å) | | Cite: | Computational redesign of PETase for plasticbiodegradation by GRAPE strategy.
Biorxiv, 2020
|
|
2FN9
 
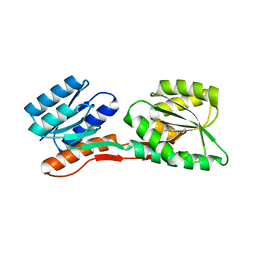 | | Thermotoga maritima Ribose Binding Protein Unliganded Form | | Descriptor: | ribose ABC transporter, periplasmic ribose-binding protein | | Authors: | Cuneo, M.J, Changela, A, Tian, Y, Beese, L.S, Hellinga, H.W. | | Deposit date: | 2006-01-10 | | Release date: | 2007-01-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Ligand-induced conformational changes in a thermophilic ribose-binding protein.
Bmc Struct.Biol., 8, 2008
|
|
5Z3V
 
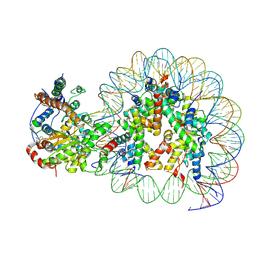 | | Structure of Snf2-nucleosome complex at shl-2 in ADP BeFx state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA (167-MER), ... | | Authors: | Li, M, Xia, X, Liu, X, Li, X, Chen, Z. | | Deposit date: | 2018-01-08 | | Release date: | 2019-05-22 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.22 Å) | | Cite: | Mechanism of DNA translocation underlying chromatin remodelling by Snf2.
Nature, 567, 2019
|
|
5Z3U
 
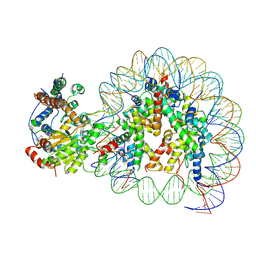 | | Structure of Snf2-nucleosome complex at shl2 in ADP BeFx state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA (167-MER), ... | | Authors: | Li, M, Xia, X, Liu, X, Li, X, Chen, Z. | | Deposit date: | 2018-01-08 | | Release date: | 2019-05-22 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.31 Å) | | Cite: | Mechanism of DNA translocation underlying chromatin remodelling by Snf2.
Nature, 567, 2019
|
|
5Z3O
 
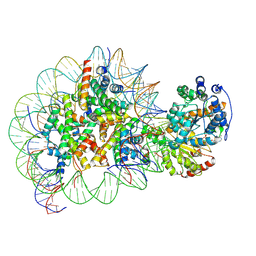 | | Structure of Snf2-nucleosome complex in ADP state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (167-MER), Histone H2A, ... | | Authors: | Li, M, Xia, X, Liu, X, Li, X, Chen, Z. | | Deposit date: | 2018-01-08 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Mechanism of DNA translocation underlying chromatin remodelling by Snf2.
Nature, 567, 2019
|
|
5Z3L
 
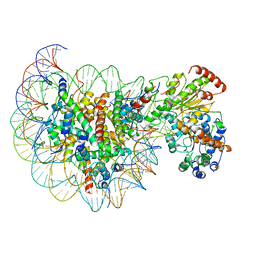 | | Structure of Snf2-nucleosome complex in apo state | | Descriptor: | DNA (167-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Li, M, Xia, X, Liu, X, Li, X, Chen, Z. | | Deposit date: | 2018-01-08 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.31 Å) | | Cite: | Mechanism of DNA translocation underlying chromatin remodelling by Snf2.
Nature, 567, 2019
|
|
7DEU
 
 | | Crystal structure of SARS-CoV-2 RBD in complex with a neutralizing antibody scFv | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody scFv | | Authors: | Zhang, Z, Zhang, G, Li, X, Rao, Z, Guo, Y. | | Deposit date: | 2020-11-05 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for SARS-CoV-2 neutralizing antibodies with novel binding epitopes.
Plos Biol., 19, 2021
|
|
7DEO
 
 | | Crystal structure of SARS-CoV-2 RBD in complex with a neutralizing antibody scFv | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Spike protein S1, ... | | Authors: | Fu, D, Zhang, G, Li, X, Rao, Z, Guo, Y. | | Deposit date: | 2020-11-04 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for SARS-CoV-2 neutralizing antibodies with novel binding epitopes.
Plos Biol., 19, 2021
|
|
7DET
 
 | | Crystal structure of SARS-CoV-2 RBD in complex with a neutralizing antibody scFv | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody scFv | | Authors: | Wang, Y, Zhang, G, Li, X, Rao, Z, Guo, Y. | | Deposit date: | 2020-11-05 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for SARS-CoV-2 neutralizing antibodies with novel binding epitopes.
Plos Biol., 19, 2021
|
|
7VWD
 
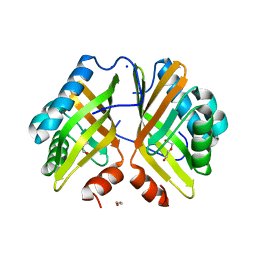 | | Crystal Structure of the Y53F/N55A mutant of LEH | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Qu, G, Li, X, Sun, Z.T, Han, X, Liu, W.D. | | Deposit date: | 2021-11-10 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.153 Å) | | Cite: | Rational enzyme design for enabling biocatalytic Baldwin cyclization and asymmetric synthesis of chiral heterocycles.
Nat Commun, 13, 2022
|
|
7VX2
 
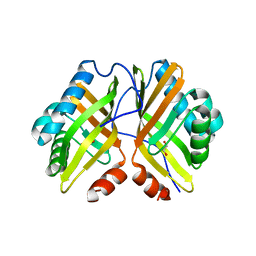 | | Crystal Structure of the Y53F/N55A/I80F/L114V/I116V mutant of LEH | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Qu, G, Li, X, Sun, Z.T, Han, X, Liu, W.D. | | Deposit date: | 2021-11-12 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.485 Å) | | Cite: | Rational enzyme design for enabling biocatalytic Baldwin cyclization and asymmetric synthesis of chiral heterocycles.
Nat Commun, 13, 2022
|
|
7VWM
 
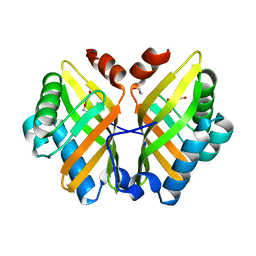 | | Crystal Structure of the Y53F/N55A/I116V mutant of LEH | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Qu, G, Li, X, Sun, Z.T, Han, X, Liu, W.D. | | Deposit date: | 2021-11-11 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Rational enzyme design for enabling biocatalytic Baldwin cyclization and asymmetric synthesis of chiral heterocycles.
Nat Commun, 13, 2022
|
|
8HQS
 
 | | Cryo-EM structure of 8-subunit Smc5/6 head region | | Descriptor: | DNA repair protein KRE29, Non-structural maintenance of chromosome element 3, Non-structural maintenance of chromosome element 4, ... | | Authors: | Qian, L, Jun, Z, Xiang, Z, Tong, C, Wang, Z, Duo, J, Zhenguo, C, Wang, L. | | Deposit date: | 2022-12-14 | | Release date: | 2024-06-19 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 31, 2024
|
|
