2KNR
 
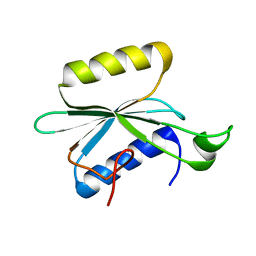 | | Solution structure of protein Atu0922 from A. tumefaciens. Northeast Structural Genomics Consortium target AtT13. Ontario Center for Structural Proteomics target ATC0905 | | Descriptor: | Uncharacterized protein ATC0905 | | Authors: | Gutmanas, A, Yee, S, Lemak, A, Fares, A, Semesi, A, Montelione, G.T, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG), Ontario Centre for Structural Proteomics (OCSP) | | Deposit date: | 2009-09-02 | | Release date: | 2009-11-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of protein atc0905 from A. tumefaciens
To be Published
|
|
3OLF
 
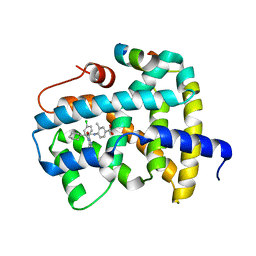 | |
3OOF
 
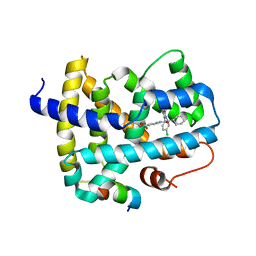 | |
3OKH
 
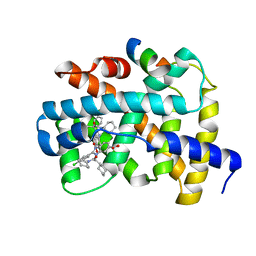 | |
3OMK
 
 | |
3OKI
 
 | |
3OOK
 
 | |
3OMM
 
 | |
4LKS
 
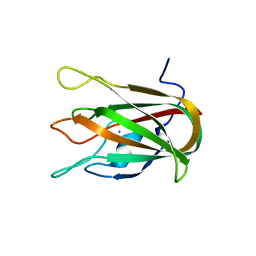 | | Structure of CBM32-3 from a family 31 glycoside hydrolase from Clostridium perfringens in complex with galactose | | Descriptor: | CALCIUM ION, Glycosyl hydrolase, family 31/fibronectin type III domain protein, ... | | Authors: | Grondin, J.M, Duan, D, Kirlin, A.C, Furness, H.S, Allingham, J.S, Smith, S.P. | | Deposit date: | 2013-07-08 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Diverse modes of galacto-specific carbohydrate recognition by a family 31 glycoside hydrolase from Clostridium perfringens.
Plos One, 12, 2017
|
|
2JOQ
 
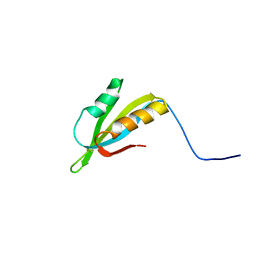 | | Solution Structure of Protein HP0495 from H. pylori; Northeast structural genomics consortium target PT2; Ontario Centre for Structural Proteomics target HP0488 | | Descriptor: | Hypothetical protein HP_0495 | | Authors: | Gutmanas, A, Lemak, A, Yee, A, Karra, M, Srisailam, S, Semesi, A, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG), Ontario Centre for Structural Proteomics (OCSP) | | Deposit date: | 2007-03-19 | | Release date: | 2007-04-03 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Protein HP0495 from H. pylori
To be Published
|
|
2JQ4
 
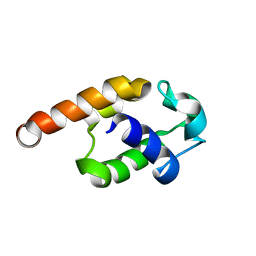 | | Complete resonance assignments and solution structure calculation of ATC2521 (NESG ID: AtT6) from Agrobacterium tumefaciens | | Descriptor: | Hypothetical protein Atu2571 | | Authors: | Srisailam, S, Lemak, A, Yee, A, Karra, M.D, Lukin, J.A, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-05-29 | | Release date: | 2007-06-12 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a protein (ATC2521)of unknown function from Agrobacterium tumefaciens
To be Published
|
|
4LPL
 
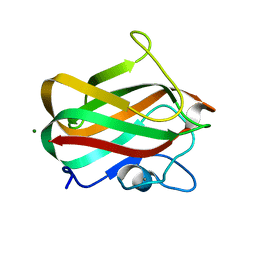 | | Structure of CBM32-1 from a family 31 glycoside hydrolase from Clostridium perfringens | | Descriptor: | CALCIUM ION, Glycosyl hydrolase, family 31/fibronectin type III domain protein, ... | | Authors: | Grondin, J.M, Duan, D, Heather, F.S, Spencer, C.A, Allingham, J.S, Smith, S.P. | | Deposit date: | 2013-07-16 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Diverse modes of galacto-specific carbohydrate recognition by a family 31 glycoside hydrolase from Clostridium perfringens.
Plos One, 12, 2017
|
|
2JN4
 
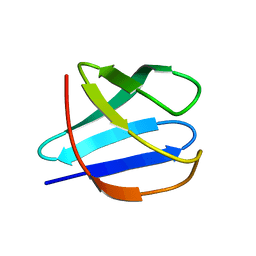 | | Solution NMR Structure of Protein RP4601 from Rhodopseudomonas palustris. Northeast Structural Genomics Consortium Target RpT2; Ontario Center for Structural Proteomics Target RP4601. | | Descriptor: | Hypothetical protein fixU, nifT | | Authors: | Lemak, A, Srisailam, S, Yee, A, Karra, M.D, Lukin, J.A, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-12-22 | | Release date: | 2007-01-23 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a hypothetical protein RP4601 from Rhodopseudomonas palustris
To be Published
|
|
4LQR
 
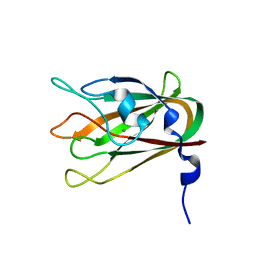 | | Structure of CBM32-3 from a family 31 glycoside hydrolase from Clostridium perfringens | | Descriptor: | CALCIUM ION, Glycosyl hydrolase, family 31/fibronectin type III domain protein | | Authors: | Grondin, J.M, Furness, H.S, Duan, D, Spencer, C.A, Allingham, J.S, Smith, S.P. | | Deposit date: | 2013-07-19 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Diverse modes of galacto-specific carbohydrate recognition by a family 31 glycoside hydrolase from Clostridium perfringens.
Plos One, 12, 2017
|
|
2JTV
 
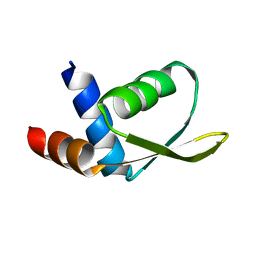 | | Solution Structure of protein RPA3401, Northeast Structural Genomics Consortium Target RpT7, Ontario Center for Structural Proteomics Target RP3384 | | Descriptor: | Protein of Unknown Function | | Authors: | Ignatchenko, A, Gutmanas, A, Lemak, A, Yee, A, Karra, M, Srisailam, S, Arrowsmith, C, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-08-07 | | Release date: | 2007-10-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of protein RPA3401, Northeast Structural Genomics Consortium Target RpT7, Ontario Center for Structural Proteomics Target RP3384
TO BE PUBLISHED
|
|
2KEO
 
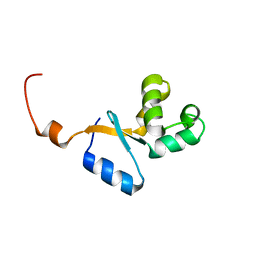 | | Solution NMR structure of human protein HS00059, cytochrome-b5-like domain of the HERC2 E3 ligase. Northeast structural genomics consortium (NESG) target ht98a | | Descriptor: | Probable E3 ubiquitin-protein ligase HERC2 | | Authors: | Lemak, A, Gutmanas, A, Fares, C, Quyang, H, Li, Y, Montelione, G, Arrowsmith, C, Dhe-Paganon, S, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-01-30 | | Release date: | 2009-02-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of human protein HS00059
To be Published
|
|
2KDB
 
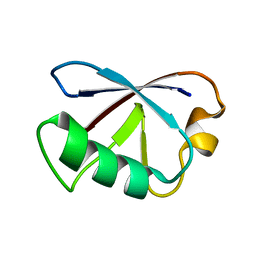 | | Solution Structure of human ubiquitin-like domain of Herpud2_9_85, Northeast Structural Genomics Consortium (NESG) target HT53A | | Descriptor: | Homocysteine-responsive endoplasmic reticulum-resident ubiquitin-like domain member 2 protein | | Authors: | Wu, B, Yee, A, Fares, C, Lemak, A, Gutmanas, A, Doherty, R, Semesi, A, Dhe-Paganon, S, Arrowsmith, C, Northeast Structural Genomics Consortium (NESG), Structural Genomics Consortium (SGC) | | Deposit date: | 2009-01-06 | | Release date: | 2009-02-10 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of human homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 2 (Herpud2 or Herp)
To be Published
|
|
6XR4
 
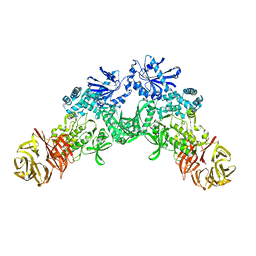 | |
4P5Y
 
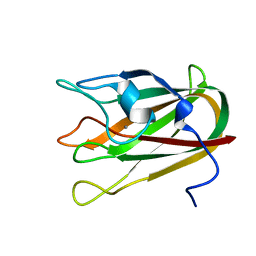 | | Structure of CBM32-3 from a family 31 glycoside hydrolase from Clostridium perfringens in complex with N-acetylgalactosamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, CALCIUM ION, Glycosyl hydrolase, ... | | Authors: | Grondin, J.M, Allingham, J.S, Boraston, A.B, Smith, S.P. | | Deposit date: | 2014-03-04 | | Release date: | 2015-10-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Diverse modes of galacto-specific carbohydrate recognition by a family 31 glycoside hydrolase from Clostridium perfringens.
PLoS ONE, 12, 2017
|
|
3C9J
 
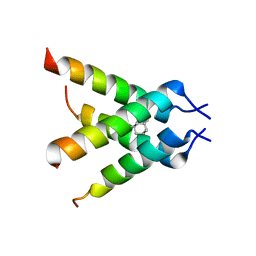 | | The Crystal structure of Transmembrane domain of M2 protein and Amantadine complex | | Descriptor: | (3S,5S,7S)-tricyclo[3.3.1.1~3,7~]decan-1-amine, Proton Channel protein M2, transmembrane segment | | Authors: | Stouffer, A.L, Acharya, R, Salom, D. | | Deposit date: | 2008-02-15 | | Release date: | 2008-03-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis for the function and inhibition of an influenza virus proton channel
Nature, 451, 2008
|
|
3CJJ
 
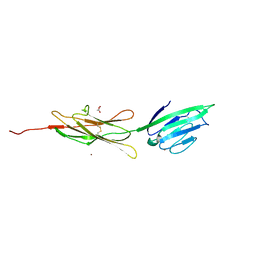 | | Crystal structure of human rage ligand-binding domain | | Descriptor: | ACETATE ION, Advanced glycosylation end product-specific receptor, ZINC ION | | Authors: | Koch, M, Dattilo, B.M, Schiefner, A, Diez, J, Chazin, W.J, Fritz, G. | | Deposit date: | 2008-03-13 | | Release date: | 2009-03-24 | | Last modified: | 2011-12-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for ligand recognition and activation of RAGE.
Structure, 18, 2010
|
|
6ZB0
 
 | |
6Z8P
 
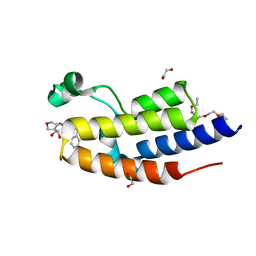 | | C-TERMINAL BROMODOMAIN OF HUMAN BRD2 WITH GSK973 | | Descriptor: | (2~{S},3~{S})-2-(fluoranylmethyl)-~{N}7-methyl-~{N}5-[(1~{R},5~{S})-3-oxabicyclo[3.1.0]hexan-6-yl]-3-phenyl-2,3-dihydro-1-benzofuran-5,7-dicarboxamide, 1,2-ETHANEDIOL, 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, ... | | Authors: | Chung, C. | | Deposit date: | 2020-06-02 | | Release date: | 2020-10-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | GSK973 Is an Inhibitor of the Second Bromodomains (BD2s) of the Bromodomain and Extra-Terminal (BET) Family.
Acs Med.Chem.Lett., 11, 2020
|
|
6ZB1
 
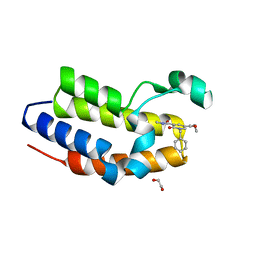 | | C-TERMINAL BROMODOMAIN OF HUMAN BRD2 WITH GSK620 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 2, ~{N}5-cyclopropyl-~{N}3-methyl-2-oxidanylidene-1-(phenylmethyl)pyridine-3,5-dicarboxamide | | Authors: | Chung, C. | | Deposit date: | 2020-06-06 | | Release date: | 2020-08-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Optimization of a Novel, Weak Bromo and Extra Terminal Domain (BET) Bromodomain Fragment Ligand to a Potent and Selective Second Bromodomain (BD2) Inhibitor.
J.Med.Chem., 63, 2020
|
|
2KKU
 
 | | Solution structure of protein af2351 from Archaeoglobus fulgidus. Northeast Structural Genomics Consortium target AtT9/Ontario Center for Structural Proteomics Target af2351 | | Descriptor: | Uncharacterized protein | | Authors: | Wu, B, Yee, A, Fares, C, Lemak, A, Rumpel, S, Semest, A, Montelione, G.T, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG), Ontario Centre for Structural Proteomics (OCSP) | | Deposit date: | 2009-06-29 | | Release date: | 2009-07-14 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution structure of protein af2351 from Archaeoglobus fulgidus. Northeast Structural Genomics Consortium target AtT9/Ontario Center for Structural Proteomics Target af2351
To be Published
|
|
