3LHP
 
 | |
3LH2
 
 | |
3TF6
 
 | |
3T1D
 
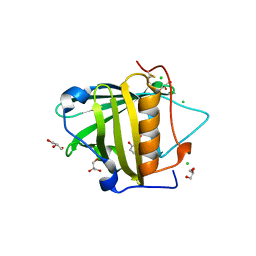 | | The mutant structure of human Siderocalin W79A, R81A, Y106F bound to Enterobactin | | Descriptor: | 2,3-DIHYDROXY-BENZOIC ACID, 2-(2,3-DIHYDROXY-BENZOYLAMINO)-3-HYDROXY-PROPIONIC ACID, CHLORIDE ION, ... | | Authors: | Seattle Structural Genomics Center for Infectious Disease, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2011-07-21 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Parsing the functional specificity of Siderocalin/Lipocalin 2/NGAL for siderophores and related small-molecule ligands.
J Struct Biol X, 2, 2019
|
|
3RPT
 
 | |
3TZS
 
 | |
3TNY
 
 | | Structure of YfiY from Bacillus cereus bound to the siderophore iron (III) schizokinen | | Descriptor: | YfiY (ABC transport system substrate-binding protein), [2-(hydroxy-kappaO)-4-[(3-{(hydroxy-kappaO)[1-(hydroxy-kappaO)ethenyl]amino}propyl)amino]-2-{2-[(3-{(hydroxy-kappaO)[1- (hydroxy-kappaO)ethenyl]amino}propyl)amino]-2-oxoethyl}-4-oxobutanoato(6-)-kappaO]iron | | Authors: | Clifton, M.C. | | Deposit date: | 2011-09-02 | | Release date: | 2012-09-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Parsing the functional specificity of Siderocalin / Lipocalin 2 / NGAL for siderophores and related small-molecule ligands
To be Published
|
|
3U0D
 
 | |
3H3P
 
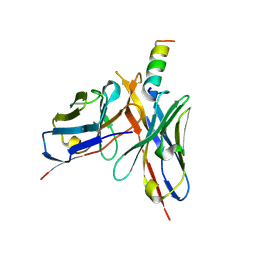 | | Crystal structure of HIV epitope-scaffold 4E10 Fv complex | | Descriptor: | 4E10_S0_1TJLC_004_N, CALCIUM ION, Fv 4E10 heavy chain, ... | | Authors: | Holmes, M.A. | | Deposit date: | 2009-04-16 | | Release date: | 2010-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Interactions between lipids and human anti-HIV antibody 4E10 can be reduced without ablating neutralizing activity
J.Virol., 84, 2010
|
|
1IGJ
 
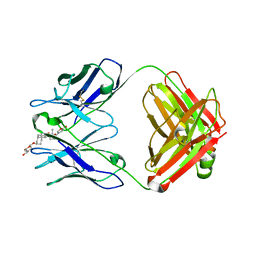 | |
1IGI
 
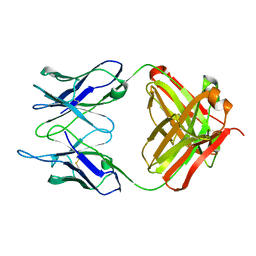 | |
7SQP
 
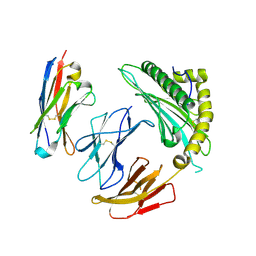 | |
7SRK
 
 | |
7SR4
 
 | |
7ST3
 
 | |
7STG
 
 | |
7SR0
 
 | | Single chain trimer HLA-A*02:01 (H98L, Y108C) with HPV.16 E7 peptide YMLDLQPET | | Descriptor: | PHOSPHATE ION, Protein E7 peptide,Beta-2-microglobulin,MHC class I antigen chimera, VHH | | Authors: | Finton, K.A.K, Rupert, P.B. | | Deposit date: | 2021-11-07 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Effects of HLA single chain trimer design on peptide presentation and stability.
Front Immunol, 14, 2023
|
|
7SR3
 
 | |
7SR5
 
 | |
7SSH
 
 | |
7U62
 
 | | Crystal structure of Anti-Heroin Antibody HY4-1F9 Fab Complexed with Morphine | | Descriptor: | (7R,7AS,12BS)-3-METHYL-2,3,4,4A,7,7A-HEXAHYDRO-1H-4,12-METHANO[1]BENZOFURO[3,2-E]ISOQUINOLINE-7,9-DIOL, ACETATE ION, HY4-1F9 Fab Heavy Chain, ... | | Authors: | Rodarte, J.V, Pancera, M.P. | | Deposit date: | 2022-03-03 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structures of drug-specific monoclonal antibodies bound to opioids and nicotine reveal a common mode of binding.
Structure, 31, 2023
|
|
7U61
 
 | | Crystal Structure of Anti-Nicotine Antibody NIC311 Fab Complexed with Nicotine | | Descriptor: | (S)-3-(1-METHYLPYRROLIDIN-2-YL)PYRIDINE, NIC311 Fab Heavy Chain, NIC311 Fab Light Chain, ... | | Authors: | Rodarte, J.V, Pancera, M.P, Liban, T.L. | | Deposit date: | 2022-03-03 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of drug-specific monoclonal antibodies bound to opioids and nicotine reveal a common mode of binding.
Structure, 31, 2023
|
|
7U64
 
 | |
6AY7
 
 | | Cartilage homing cysteine-dense-peptides | | Descriptor: | CHLORIDE ION, Potassium channel toxin alpha-KTx 3.5, SULFATE ION, ... | | Authors: | Gewe, M.M, Strong, R.K. | | Deposit date: | 2017-09-07 | | Release date: | 2018-08-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Cartilage homing cysteine-dense-peptides
To Be Published
|
|
6ATY
 
 | |
