6LUD
 
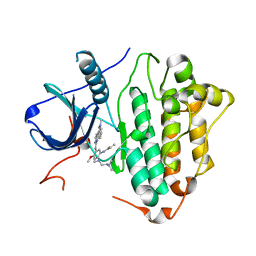 | | Crystal Structure of EGFR(L858R/T790M/C797S) in complex with Osimertinib | | Descriptor: | Epidermal growth factor receptor, N-(2-{[2-(dimethylamino)ethyl](methyl)amino}-4-methoxy-5-{[4-(1-methyl-1H-indol-3-yl)pyrimidin-2-yl]amino}phenyl)prop-2-enamide | | Authors: | Kawauchi, H, Fukami, T.A, Sato, S, Endo, M, Torizawa, T, Kashima, K, Chiba, T, Sakamoto, H. | | Deposit date: | 2020-01-27 | | Release date: | 2020-10-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | CH7233163 Overcomes Osimertinib-Resistant EGFR-Del19/T790M/C797S Mutation.
Mol.Cancer Ther., 19, 2020
|
|
2BTG
 
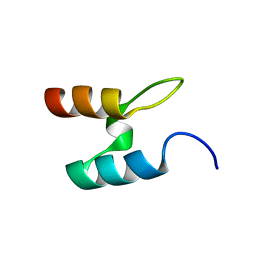 | |
2BTH
 
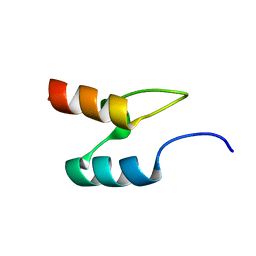 | |
1M3B
 
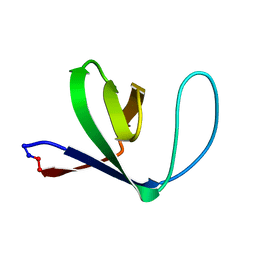 | | Solution structure of a circular form of the N-terminal SH3 domain (A134C, E135G, R191G mutant) from oncogene protein c-Crk. | | Descriptor: | Proto-oncogene C-crk | | Authors: | Schumann, F.H, Varadan, R, Tayakuniyil, P.P, Hall, J.B, Camarero, J.A, Fushman, D. | | Deposit date: | 2002-06-27 | | Release date: | 2003-08-05 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Changing protein backbone topology: Structural and dynamic consequences of the backbone cyclization in SH3 domain
To be Published
|
|
1M3A
 
 | | Solution structure of a circular form of the truncated N-terminal SH3 domain from oncogene protein c-Crk. | | Descriptor: | Proto-oncogene C-crk | | Authors: | Schumann, F.H, Varadan, R, Tayakuniyil, P.P, Hall, J.B, Camarero, J.A, Fushman, D. | | Deposit date: | 2002-06-27 | | Release date: | 2003-08-05 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Changing protein backbone topology: Structural and dynamic consequences of the backbone cyclization in SH3 domain
To be Published
|
|
1M30
 
 | | Solution structure of N-terminal SH3 domain from oncogene protein c-Crk | | Descriptor: | Proto-oncogene C-crk | | Authors: | Schumann, F.H, Varadan, R, Tayakuniyil, P.P, Hall, J.B, Camarero, J.A, Fushman, D. | | Deposit date: | 2002-06-26 | | Release date: | 2003-08-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Changing protein backbone topology: Structural and dynamic consequences of the backbone cyclization in SH3 domain
To be Published
|
|
1M3C
 
 | | Solution structure of a circular form of the N-terminal SH3 domain (E132C, E133G, R191G mutant) from oncogene protein c-Crk | | Descriptor: | Proto-oncogene C-crk | | Authors: | Schumann, F.H, Varadan, R, Tayakuniyil, P.P, Hall, J.B, Camarero, J.A, Fushman, D. | | Deposit date: | 2002-06-27 | | Release date: | 2003-08-05 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Changing protein backbone topology: Structural and dynamic consequences of the backbone cyclization in SH3 domain
To be Published
|
|
2ELL
 
 | | Solution structure of the Leucine Rich Repeat of human Acidic leucine-rich nuclear phosphoprotein 32 family member B | | Descriptor: | Acidic leucine-rich nuclear phosphoprotein 32 family member B | | Authors: | Tochio, N, Koshiba, S, Watanabe, S, Harada, T, Umehara, T, Tanaka, A, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-27 | | Release date: | 2008-04-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of histone chaperone ANP32B: interaction with core histones H3-H4 through its acidic concave domain.
J.Mol.Biol., 401, 2010
|
|
2EQA
 
 | | Crystal Structure of the hypothetical Sua5 protein from Sulfolobus tokodaii | | Descriptor: | ADENOSINE MONOPHOSPHATE, Hypothetical protein ST1526, MAGNESIUM ION | | Authors: | Agari, Y, Shinkai, A, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-30 | | Release date: | 2008-01-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystal structure of a hypothetical Sua5 protein from Sulfolobus tokodaii strain 7
Proteins, 70, 2008
|
|
3VHA
 
 | | Hsp90 alpha N-terminal domain in complex with a macrocyclic inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 22-methyl-13,18-dioxa-7-thia-3,5-diazatetracyclo[17.3.1.1~2,6~.1~8,12~]pentacosa-1(23),2(25),3,5,8(24),9,11,19,21-nonaen-4-amine, Heat shock protein HSP 90-alpha | | Authors: | Fukami, T.A, Ono, N. | | Deposit date: | 2011-08-24 | | Release date: | 2012-07-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Design and synthesis of novel macrocyclic 2-amino-6-arylpyrimidine Hsp90 inhibitors
Bioorg.Med.Chem.Lett., 22, 2012
|
|
3VHC
 
 | | Hsp90 alpha N-terminal domain in complex with a macrocyclic inhibitor | | Descriptor: | 4-amino-20,22-dimethyl-13-oxa-7-thia-3,5,17-triazatetracyclo[17.3.1.1~2,6~.1~8,12~]pentacosa-1(23),2(25),3,5,8(24),9,11,19,21-nonaen-18-one, Heat shock protein HSP 90-alpha, MAGNESIUM ION | | Authors: | Fukami, T.A, Ono, N. | | Deposit date: | 2011-08-24 | | Release date: | 2012-07-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Design and synthesis of novel macrocyclic 2-amino-6-arylpyrimidine Hsp90 inhibitors
Bioorg.Med.Chem.Lett., 22, 2012
|
|
3VHD
 
 | | Hsp90 alpha N-terminal domain in complex with a macrocyclic inhibitor, CH5164840 | | Descriptor: | 4-amino-18,20-dimethyl-7-thia-3,5,11,15-tetraazatricyclo[15.3.1.1(2,6)]docosa-1(20),2,4,6(22),17(21),18-hexaene-10,16-dione, Heat shock protein HSP 90-alpha | | Authors: | Fukami, T.A, Ono, N. | | Deposit date: | 2011-08-24 | | Release date: | 2012-07-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Design and synthesis of novel macrocyclic 2-amino-6-arylpyrimidine Hsp90 inhibitors
Bioorg.Med.Chem.Lett., 22, 2012
|
|
1W4H
 
 | | Peripheral-subunit from mesophilic, thermophilic and hyperthermophilic bacteria fold by ultrafast, apparently two-state transitions | | Descriptor: | DIHYDROLIPOYLLYSINE-RESIDUE ACETYLTRANSFERASE | | Authors: | Ferguson, N, Sharpe, T.D, Schartau, P.J, Allen, M.D, Johnson, C.M, Fersht, A.R. | | Deposit date: | 2004-07-23 | | Release date: | 2005-07-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Ultra-Fast Barrier-Limited Folding in the Peripheral Subunit-Binding Domain Family.
J.Mol.Biol., 353, 2005
|
|
1W4F
 
 | | Peripheral-subunit from mesophilic, thermophilic and hyperthermophilic bacteria fold by ultrafast, apparently two-state transitions | | Descriptor: | DIHYDROLIPOYLLYSINE-RESIDUE ACETYLTRANSFERASE | | Authors: | Ferguson, N, Sharpe, T.D, Schartau, P.J, Allen, M.D, Johnson, C.M, Fersht, A.R. | | Deposit date: | 2004-07-23 | | Release date: | 2005-07-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Ultra-Fast Barrier-Limited Folding in the Peripheral Subunit-Binding Domain Family.
J.Mol.Biol., 353, 2005
|
|
3A6R
 
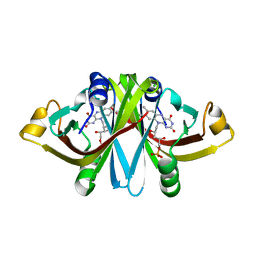 | | E13Q mutant of FMN-binding protein from Desulfovibrio vulgaris (Miyazaki F) | | Descriptor: | CHLORIDE ION, FLAVIN MONONUCLEOTIDE, FMN-binding protein | | Authors: | Nakanishi, T, Haruyama, Y, Inoue, H, Kitamura, M. | | Deposit date: | 2009-09-08 | | Release date: | 2010-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Effects of the disappearance of one charge on ultrafast fluorescence dynamics of the FMN binding protein.
J.Phys.Chem.B, 114, 2010
|
|
3A6Q
 
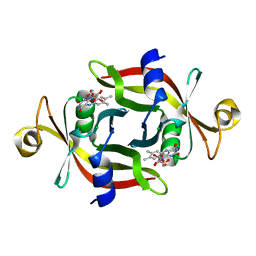 | | E13T mutant of FMN-binding protein from Desulfovibrio vulgaris (Miyazaki F) | | Descriptor: | CHLORIDE ION, FLAVIN MONONUCLEOTIDE, FMN-binding protein | | Authors: | Nakanishi, T, Haruyama, Y, Inoue, H, Kitamura, M. | | Deposit date: | 2009-09-08 | | Release date: | 2010-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Effects of the disappearance of one charge on ultrafast fluorescence dynamics of the FMN binding protein.
J.Phys.Chem.B, 114, 2010
|
|
2HBB
 
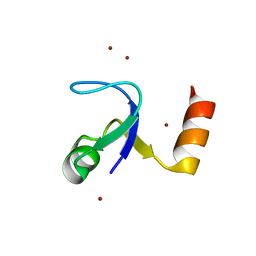 | | Crystal Structure of the N-terminal Domain of Ribosomal Protein L9 (NTL9) | | Descriptor: | 50S ribosomal protein L9, ZINC ION | | Authors: | Cho, J.-H, Kim, E.Y, Schindelin, H, Raleigh, D.P. | | Deposit date: | 2006-06-14 | | Release date: | 2007-05-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Energetically significant networks of coupled interactions within an unfolded protein.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2HBA
 
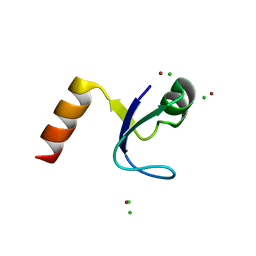 | | Crystal Structure of N-terminal Domain of Ribosomal Protein L9 (NTL9) K12M | | Descriptor: | 50S ribosomal protein L9, CHLORIDE ION, IMIDAZOLE, ... | | Authors: | Cho, J.-H, Kim, E.Y, Schindelin, H, Raleigh, D.P. | | Deposit date: | 2006-06-14 | | Release date: | 2007-05-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Energetically significant networks of coupled interactions within an unfolded protein.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1UEK
 
 | | Crystal structure of 4-(cytidine 5'-diphospho)-2C-methyl-D-erythritol kinase | | Descriptor: | 4-(cytidine 5'-diphospho)-2C-methyl-D-erythritol kinase | | Authors: | Wada, T, Kuramitsu, S, Yokoyama, S, Tame, J.R.H, Park, S.Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-05-17 | | Release date: | 2003-06-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of 4-(Cytidine 5'-diphospho)-2-C-methyl-D-erythritol kinase, an Enzyme in the Non-mevalonate Pathway of Isoprenoid Synthesis.
J.Biol.Chem., 278, 2003
|
|
2DJW
 
 | | Crystal structure of TTHA0845 from Thermus thermophilus HB8 | | Descriptor: | ZINC ION, probable transcriptional regulator, AsnC family | | Authors: | Okazaki, N, Nakano, N, Shinkai, A, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-04-05 | | Release date: | 2006-09-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the stand-alone RAM-domain protein from Thermus thermophilus HB8
ACTA CRYSTALLOGR.,SECT.F, 62, 2006
|
|
3LMP
 
 | | Crystal structure of the PPARgamma-LBD complexed with a cercosporamide derivative modulator | | Descriptor: | (9aS)-8-acetyl-1,7-dihydroxy-3-methoxy-9a-methyl-N-(1-naphthylmethyl)-9-oxo-9,9a-dihydrodibenzo[b,d]furan-4-carboxamide, Peptide of Nuclear receptor coactivator 1, Peroxisome proliferator-activated receptor gamma | | Authors: | Matsui, Y, Hanzawa, H. | | Deposit date: | 2010-01-31 | | Release date: | 2010-04-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of a novel selective PPARgamma modulator from (-)-Cercosporamide derivatives
Bioorg.Med.Chem.Lett., 20, 2010
|
|
4F9M
 
 | | Crystal structure of the PPARgamma-LBD complexed with a cercosporamide derivative modulator | | Descriptor: | (9aS)-8-acetyl-N-[(2-ethyl-4-fluoronaphthalen-1-yl)methyl]-1,7-dihydroxy-3-methoxy-9a-methyl-9-oxo-9,9a-dihydrodibenzo[b,d]furan-4-carboxamide, Peroxisome proliferator-activated receptor gamma, peptide from Nuclear receptor coactivator 1 | | Authors: | Matsui, Y, Hanzawa, H. | | Deposit date: | 2012-05-19 | | Release date: | 2012-08-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Synthesis and biological evaluation of novel (-)-cercosporamide derivatives as potent selective PPARg modulators
Eur.J.Med.Chem., 54, 2012
|
|
2CB1
 
 | | Crystal Structure of O-actetyl Homoserine Sulfhydrylase From Thermus Thermophilus HB8,OAH2. | | Descriptor: | O-ACETYL HOMOSERINE SULFHYDRYLASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Imagawa, T, Utsunomiya, H, Tsuge, H, Ebihara, A, Kanagawa, M, Nakagawa, N, Kuroishi, C, Agari, Y, Kuramitsu, S, Yokoyama, S. | | Deposit date: | 2005-12-28 | | Release date: | 2007-01-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of O-Acetyl Homoserine Sulfhydrylase
To be Published
|
|
3V9T
 
 | | Crystal structure of the PPARgamma-LBD complexed with a cercosporamide derivative modulator | | Descriptor: | (9aS)-8-acetyl-N-[(3-ethoxynaphthalen-1-yl)methyl]-1,7-dihydroxy-3-methoxy-9a-methyl-9-oxo-9,9a-dihydrodibenzo[b,d]furan-4-carboxamide, Peptide from Peroxisome proliferator-activated receptor gamma coactivator 1-alpha, Peroxisome proliferator-activated receptor gamma | | Authors: | Matsui, Y, Hanzawa, H. | | Deposit date: | 2011-12-28 | | Release date: | 2012-02-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Substituents at the naphthalene C3 position of (-)-Cercosporamide derivatives significantly affect the maximal efficacy as PPAR(gamma) partial agonists
Bioorg.Med.Chem.Lett., 22, 2012
|
|
3V9V
 
 | | Crystal structure of the PPARgamma-LBD complexed with a cercosporamide derivative modulator | | Descriptor: | Peptide from Peroxisome proliferator-activated receptor gamma coactivator 1-alpha, Peroxisome proliferator-activated receptor gamma, methyl 3-{4-[({[(9aS)-8-acetyl-1,7-dihydroxy-3-methoxy-9a-methyl-9-oxo-9,9a-dihydrodibenzo[b,d]furan-4-yl]carbonyl}amino)methyl]naphthalen-2-yl}propanoate | | Authors: | Matsui, Y, Hanzawa, H. | | Deposit date: | 2011-12-28 | | Release date: | 2012-02-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Substituents at the naphthalene C3 position of (-)-Cercosporamide derivatives significantly affect the maximal efficacy as PPAR(gamma) partial agonists
Bioorg.Med.Chem.Lett., 22, 2011
|
|
